| Gene symbol |
Time course plot |
Accession |
Description |
Treatment |
15min |
30min |
1hr |
2hr |
4hr |
8hr |
20hr |
AK053501
| 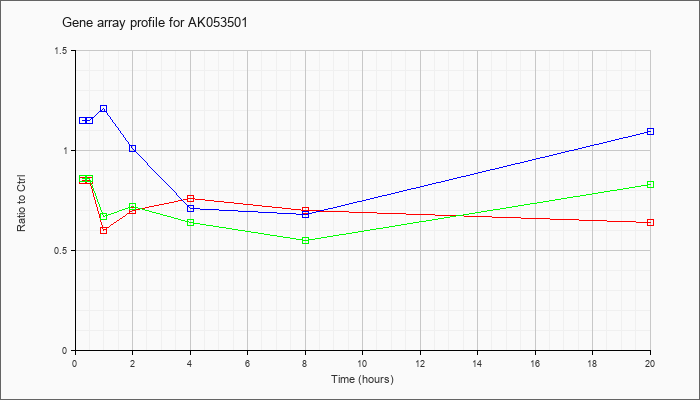 |
AK053501 |
0 day neonate eyeball cDNA, RIKEN full-length enriched library, clone:E130103H07 product:inferred: ADP-ribosylation factor-directed GTPase activating protein isoform b {Mus musculus}, full insert sequence. [AK053501] |
KLA | .85 |
.71 |
.60 |
.70 |
.76 |
.70 |
.64 |
| ATP | 1.15 |
1.25 |
1.21 |
1.01 |
.71 |
.68 |
1.09 |
| KLA/ATP | .86 |
.89 |
.67 |
.72 |
.64 |
.55 |
.83 |
|
| Akt1 | 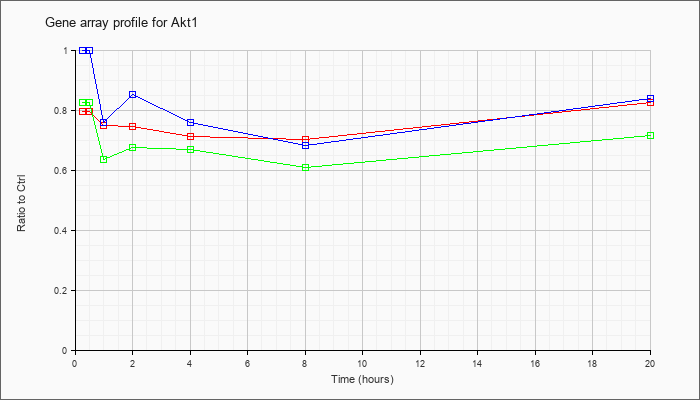 |
NM_009652 |
thymoma viral proto-oncogene 1 (Akt1), mRNA [NM_009652] |
KLA | .80 |
.80 |
.75 |
.75 |
.71 |
.70 |
.82 |
| ATP | 1.00 |
1.02 |
.76 |
.85 |
.76 |
.68 |
.84 |
| KLA/ATP | .82 |
.80 |
.64 |
.68 |
.67 |
.61 |
.72 |
|
| Akt2 | 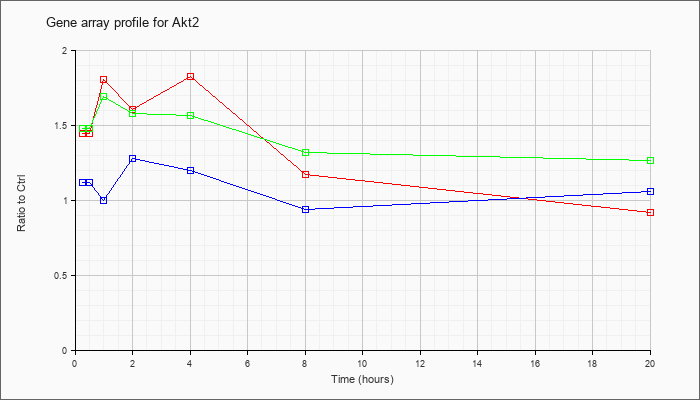 |
NM_007434 |
thymoma viral proto-oncogene 2 (Akt2), transcript variant 2, mRNA [NM_007434] |
KLA | 1.45 |
1.49 |
1.81 |
1.61 |
1.83 |
1.17 |
.92 |
| ATP | 1.12 |
1.12 |
1.00 |
1.28 |
1.20 |
.94 |
1.06 |
| KLA/ATP | 1.48 |
1.70 |
1.69 |
1.58 |
1.57 |
1.32 |
1.27 |
|
| Akt3 | 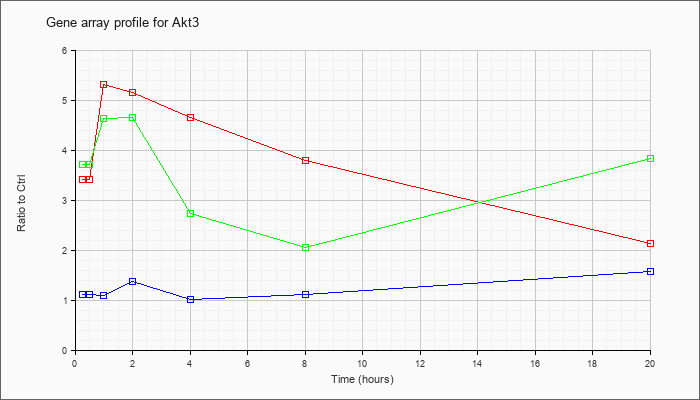 |
NM_011785 |
thymoma viral proto-oncogene 3 (Akt3), mRNA [NM_011785] |
KLA | 3.42 |
3.45 |
5.31 |
5.16 |
4.65 |
3.80 |
2.12 |
| ATP | 1.12 |
1.22 |
1.10 |
1.37 |
1.02 |
1.11 |
1.56 |
| KLA/ATP | 3.72 |
4.17 |
4.62 |
4.64 |
2.74 |
2.05 |
3.82 |
|
| Amph |  |
AK047144 |
10 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:B930026D14 product:similar to MYC BOX DEPENDENT INTERACTING PROTEIN 1 (BRIDGING INTEGRATOR 1) (AMPHIPHYSIN-LIKE PROTEIN) (AMPHIPHYSIN II) [Rattus norvegicus], full |
KLA | .96 |
.98 |
.94 |
.99 |
1.06 |
1.02 |
1.04 |
| ATP | 1.00 |
1.02 |
1.06 |
.99 |
.98 |
1.06 |
1.00 |
| KLA/ATP | .97 |
.94 |
1.08 |
1.02 |
1.00 |
1.04 |
.97 |
|
| Amph |  |
NM_175007 |
amphiphysin (Amph), mRNA [NM_175007] |
KLA | .97 |
1.04 |
1.04 |
1.04 |
1.04 |
.99 |
1.02 |
| ATP | 1.01 |
1.02 |
1.00 |
.98 |
.95 |
1.01 |
1.02 |
| KLA/ATP | 1.00 |
1.08 |
.96 |
1.04 |
.97 |
1.00 |
.94 |
|
| Arf6 |  |
NM_007481 |
ADP-ribosylation factor 6 (Arf6), mRNA [NM_007481] |
KLA | .99 |
1.01 |
1.19 |
1.22 |
1.41 |
1.33 |
1.13 |
| ATP | 1.02 |
1.12 |
1.06 |
1.43 |
1.16 |
1.12 |
1.37 |
| KLA/ATP | 1.04 |
1.01 |
.91 |
1.32 |
1.16 |
1.32 |
1.52 |
|
| Arpc1a |  |
NM_019767 |
actin related protein 2/3 complex, subunit 1A (Arpc1a), mRNA [NM_019767] |
KLA | .80 |
.78 |
.73 |
.79 |
.90 |
1.02 |
.97 |
| ATP | 1.01 |
1.07 |
.93 |
.86 |
.83 |
1.14 |
1.15 |
| KLA/ATP | .77 |
.74 |
.64 |
.64 |
.77 |
1.13 |
1.51 |
|
| Arpc1b |  |
NM_023142 |
actin related protein 2/3 complex, subunit 1B (Arpc1b), mRNA [NM_023142] |
KLA | .85 |
.86 |
.81 |
.83 |
.92 |
.93 |
1.03 |
| ATP | 1.03 |
.98 |
.86 |
.79 |
.84 |
.85 |
.91 |
| KLA/ATP | .88 |
.84 |
.83 |
.73 |
.71 |
.70 |
.63 |
|
| Arpc2 | 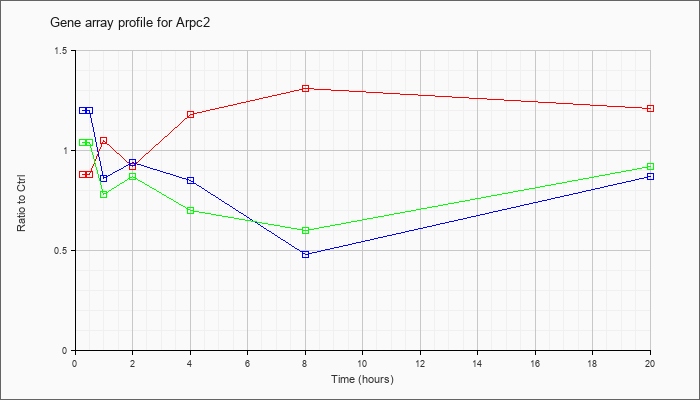 |
NM_029711 |
actin related protein 2/3 complex, subunit 2 (Arpc2), mRNA [NM_029711] |
KLA | .88 |
.89 |
1.05 |
.92 |
1.18 |
1.31 |
1.21 |
| ATP | 1.20 |
1.18 |
.86 |
.94 |
.85 |
.48 |
.87 |
| KLA/ATP | 1.04 |
1.05 |
.78 |
.87 |
.70 |
.60 |
.92 |
|
| Arpc3 | 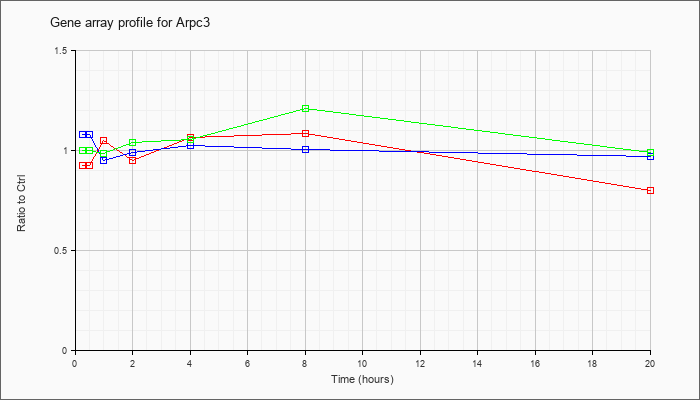 |
NM_019824 |
actin related protein 2/3 complex, subunit 3 (Arpc3), mRNA [NM_019824] |
KLA | .93 |
.96 |
1.05 |
.95 |
1.07 |
1.09 |
.80 |
| ATP | 1.08 |
1.05 |
.95 |
.99 |
1.03 |
1.01 |
.97 |
| KLA/ATP | 1.00 |
1.02 |
.99 |
1.04 |
1.06 |
1.21 |
.99 |
|
| Arpc4 | 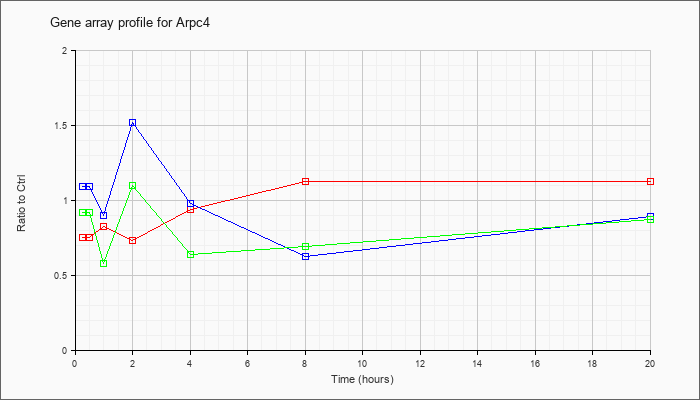 |
NM_026552 |
actin related protein 2/3 complex, subunit 4 (Arpc4), mRNA [NM_026552] |
KLA | .75 |
.81 |
.83 |
.73 |
.94 |
1.12 |
1.12 |
| ATP | 1.09 |
1.34 |
.90 |
1.52 |
.98 |
.63 |
.89 |
| KLA/ATP | .92 |
.90 |
.58 |
1.10 |
.64 |
.69 |
.87 |
|
| Arpc5 | 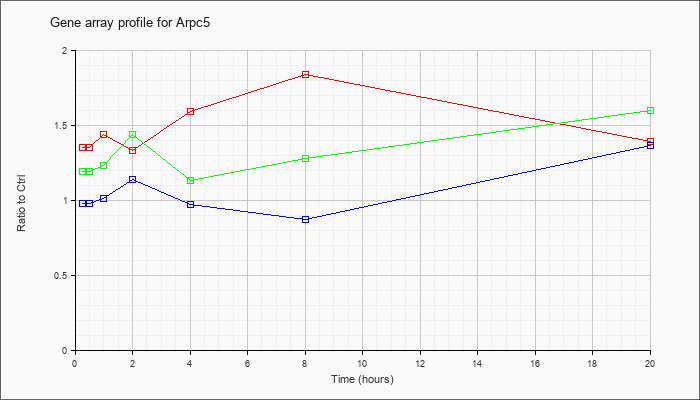 |
NM_026369 |
actin related protein 2/3 complex, subunit 5 (Arpc5), mRNA [NM_026369] |
KLA | 1.35 |
1.37 |
1.44 |
1.33 |
1.59 |
1.84 |
1.39 |
| ATP | .98 |
1.08 |
1.01 |
1.14 |
.97 |
.87 |
1.36 |
| KLA/ATP | 1.19 |
1.30 |
1.23 |
1.44 |
1.13 |
1.28 |
1.60 |
|
| Arpc5l | 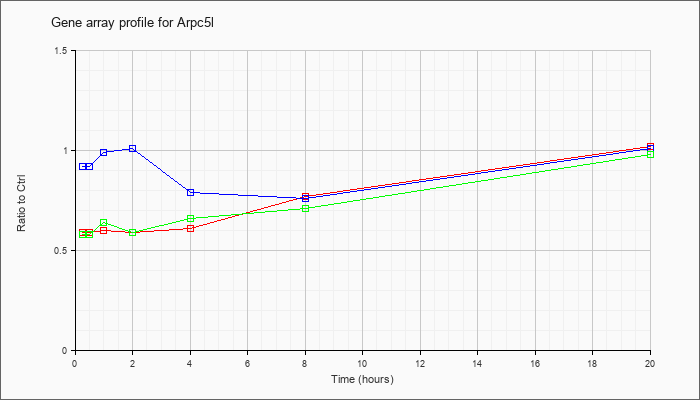 |
NM_028809 |
actin related protein 2/3 complex, subunit 5-like (Arpc5l), mRNA [NM_028809] |
KLA | .59 |
.61 |
.60 |
.59 |
.61 |
.77 |
1.02 |
| ATP | .92 |
.92 |
.99 |
1.01 |
.79 |
.76 |
1.01 |
| KLA/ATP | .58 |
.60 |
.64 |
.59 |
.66 |
.71 |
.98 |
|
| Cdc42 | 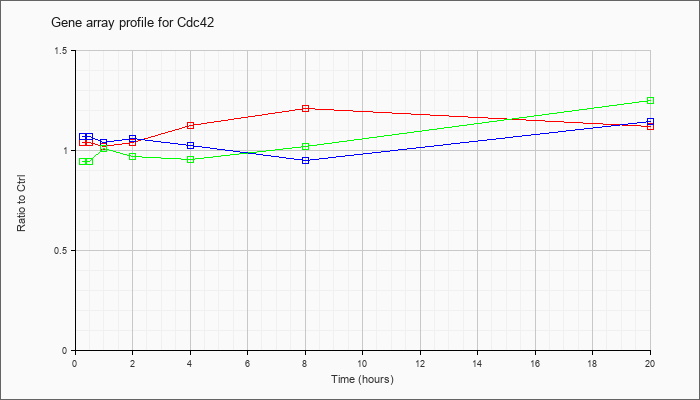 |
NM_009861 |
cell division cycle 42 homolog (S. cerevisiae) (Cdc42), mRNA [NM_009861] |
KLA | 1.04 |
1.01 |
1.02 |
1.04 |
1.12 |
1.21 |
1.12 |
| ATP | 1.07 |
1.10 |
1.04 |
1.06 |
1.02 |
.95 |
1.14 |
| KLA/ATP | .94 |
1.04 |
1.01 |
.97 |
.95 |
1.02 |
1.25 |
|
| Cfl1 | 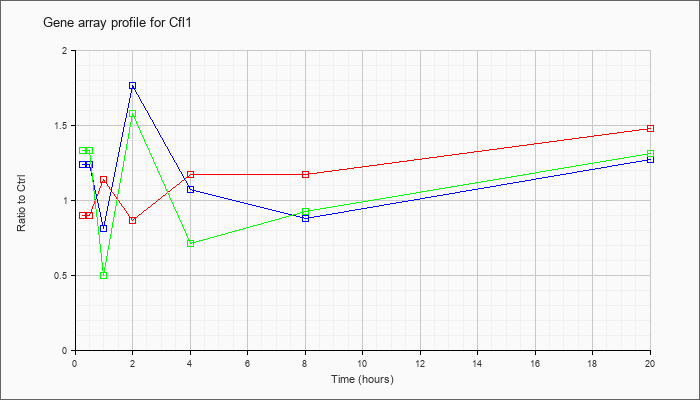 |
NM_007687 |
cofilin 1, non-muscle (Cfl1), mRNA [NM_007687] |
KLA | .90 |
1.01 |
1.14 |
.87 |
1.17 |
1.17 |
1.48 |
| ATP | 1.24 |
1.60 |
.81 |
1.77 |
1.07 |
.88 |
1.27 |
| KLA/ATP | 1.33 |
1.48 |
.50 |
1.58 |
.71 |
.93 |
1.31 |
|
| Cfl2 | 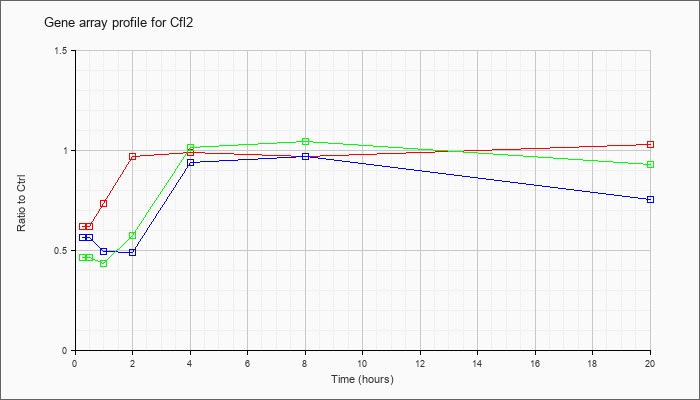 |
NM_007688 |
cofilin 2, muscle (Cfl2), mRNA [NM_007688] |
KLA | .62 |
.65 |
.74 |
.97 |
.99 |
.97 |
1.03 |
| ATP | .57 |
.49 |
.50 |
.49 |
.94 |
.97 |
.76 |
| KLA/ATP | .47 |
.41 |
.44 |
.58 |
1.02 |
1.05 |
.93 |
|
| Crk | 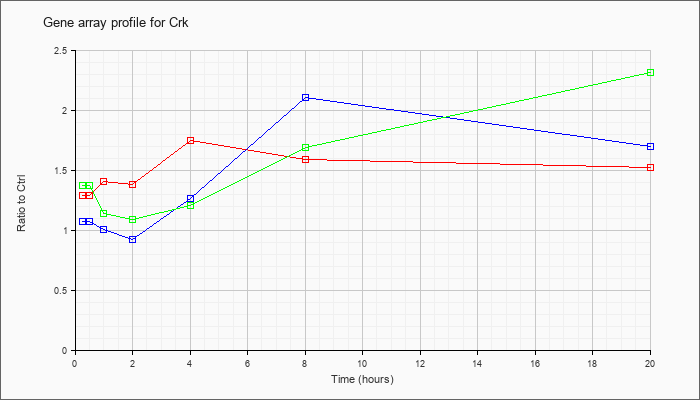 |
NM_133656 |
v-crk sarcoma virus CT10 oncogene homolog (avian) (Crk), mRNA [NM_133656] |
KLA | 1.29 |
1.32 |
1.41 |
1.38 |
1.75 |
1.59 |
1.52 |
| ATP | 1.08 |
1.17 |
1.01 |
.93 |
1.27 |
2.11 |
1.70 |
| KLA/ATP | 1.37 |
1.49 |
1.14 |
1.09 |
1.21 |
1.69 |
2.32 |
|
| Crkl | 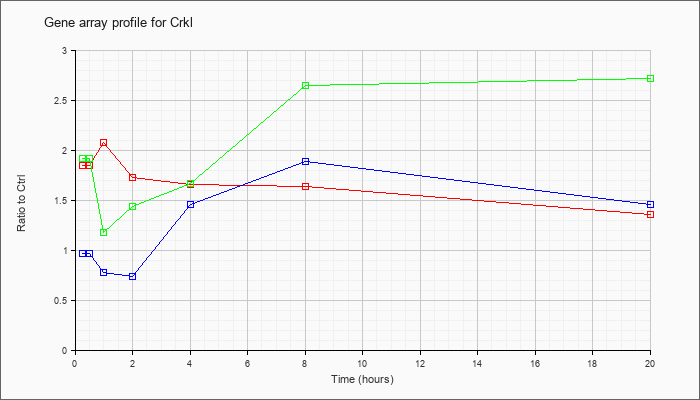 |
NM_007764 |
v-crk sarcoma virus CT10 oncogene homolog (avian)-like (Crkl), mRNA [NM_007764] |
KLA | 1.85 |
1.97 |
2.08 |
1.73 |
1.66 |
1.64 |
1.36 |
| ATP | .97 |
.94 |
.78 |
.74 |
1.46 |
1.89 |
1.46 |
| KLA/ATP | 1.92 |
1.88 |
1.18 |
1.44 |
1.67 |
2.65 |
2.72 |
|
| Ddef1 | 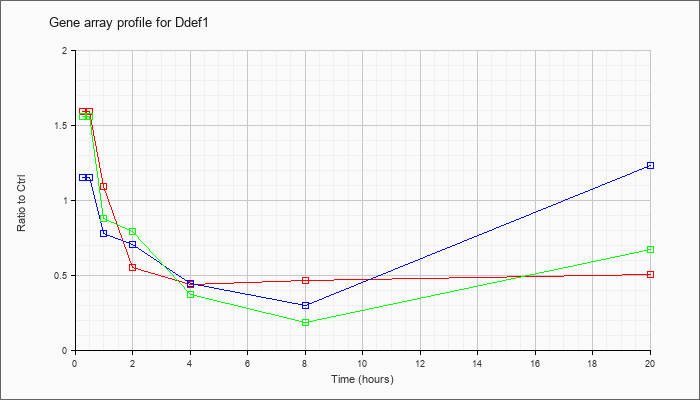 |
AK147689 |
14 days pregnant adult female placenta cDNA, RIKEN full-length enriched library, clone:M5C1108F01 product:development and differentiation enhancing, full insert sequence [AK147689] |
KLA | 1.39 |
1.17 |
.77 |
.40 |
.35 |
.37 |
.43 |
| ATP | 1.06 |
1.02 |
.78 |
.58 |
.32 |
.20 |
.83 |
| KLA/ATP | 1.17 |
1.12 |
.69 |
.57 |
.21 |
.13 |
.29 |
|
| Ddef1 |  |
NM_010026 |
development and differentiation enhancing (Ddef1), mRNA [NM_010026] |
KLA | 1.79 |
1.52 |
1.41 |
.70 |
.52 |
.56 |
.58 |
| ATP | 1.24 |
1.13 |
.78 |
.83 |
.57 |
.40 |
1.63 |
| KLA/ATP | 1.95 |
1.71 |
1.07 |
1.01 |
.53 |
.24 |
1.05 |
|
| Ddef2 | 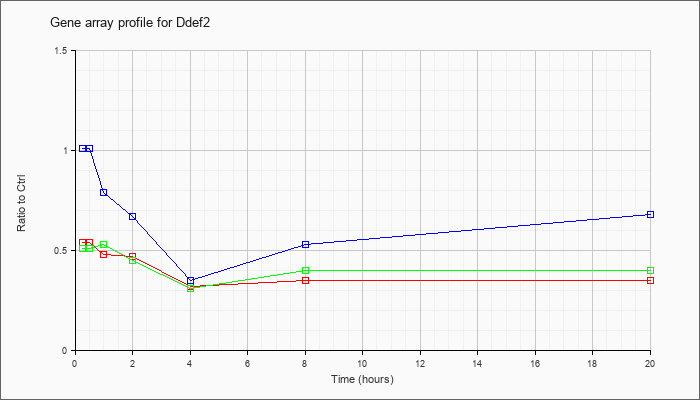 |
NM_001004364 |
development and differentiation enhancing factor 2 (Ddef2), transcript variant 2, mRNA [NM_001004364] |
KLA | .54 |
.52 |
.48 |
.47 |
.32 |
.35 |
.35 |
| ATP | 1.01 |
1.05 |
.79 |
.67 |
.35 |
.53 |
.68 |
| KLA/ATP | .51 |
.52 |
.53 |
.45 |
.31 |
.40 |
.40 |
|
| Ddefl1 | 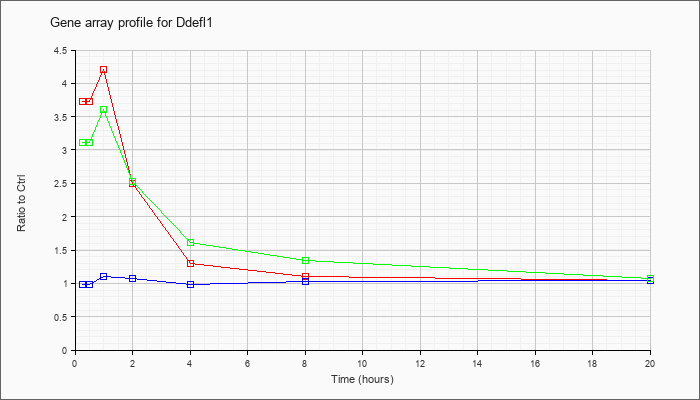 |
AK035099 |
12 days embryo embryonic body between diaphragm region and neck cDNA, RIKEN full-length enriched library, clone:9430088F20 product:hypothetical Ankyrin repeat profile/Serine-rich region/Ankyrin-repeat/Ankyrin repeat region circular profil |
KLA | 3.72 |
3.96 |
4.21 |
2.49 |
1.30 |
1.10 |
1.05 |
| ATP | .99 |
.94 |
1.10 |
1.07 |
.99 |
1.03 |
1.04 |
| KLA/ATP | 3.11 |
4.46 |
3.60 |
2.53 |
1.62 |
1.34 |
1.08 |
|
| Dnm2 | 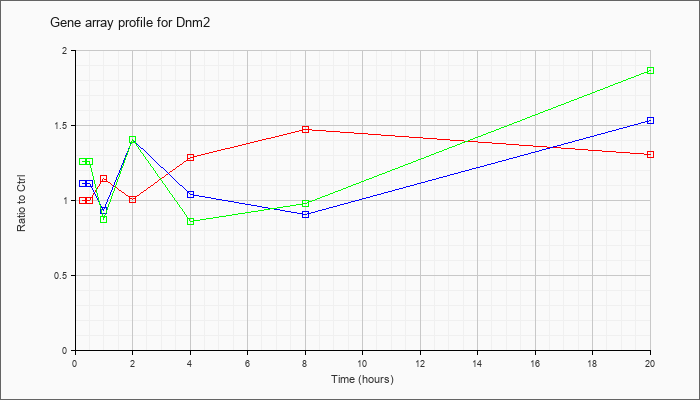 |
NM_001039520 |
dynamin 2 (Dnm2), mRNA [NM_001039520] |
KLA | 1.00 |
1.03 |
1.14 |
1.01 |
1.29 |
1.47 |
1.30 |
| ATP | 1.11 |
1.31 |
.93 |
1.41 |
1.04 |
.90 |
1.53 |
| KLA/ATP | 1.26 |
1.23 |
.87 |
1.41 |
.86 |
.98 |
1.86 |
|
| Dock2 | 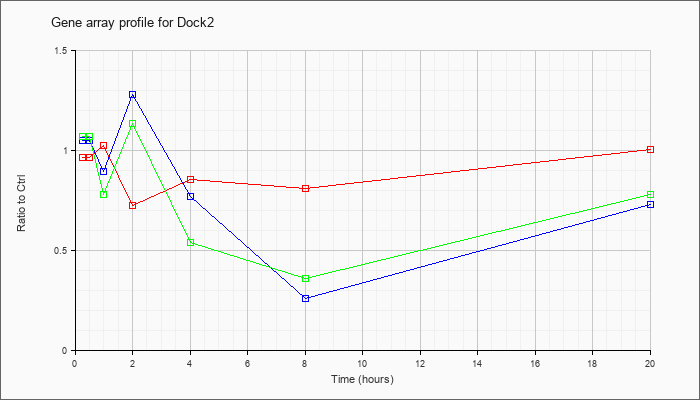 |
NM_033374 |
dedicator of cyto-kinesis 2 (Dock2), mRNA [NM_033374] |
KLA | .97 |
.95 |
1.03 |
.73 |
.86 |
.81 |
1.01 |
| ATP | 1.05 |
1.18 |
.90 |
1.28 |
.77 |
.26 |
.73 |
| KLA/ATP | 1.07 |
1.12 |
.78 |
1.14 |
.54 |
.36 |
.78 |
|
EG629860
| 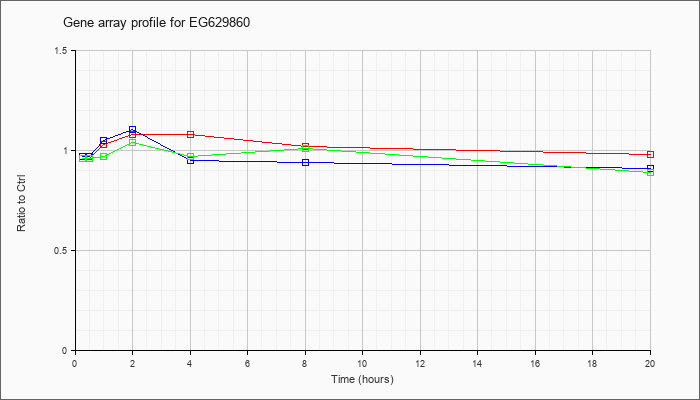 |
M17953 |
gb|Mouse Ig rearranged H-chain V-region mRNA VJ1 [M17953] |
KLA | .96 |
1.05 |
1.03 |
1.08 |
1.08 |
1.02 |
.98 |
| ATP | .97 |
.99 |
1.05 |
1.10 |
.95 |
.94 |
.91 |
| KLA/ATP | .96 |
.94 |
.97 |
1.04 |
.97 |
1.01 |
.89 |
|
| Fcgr1 | 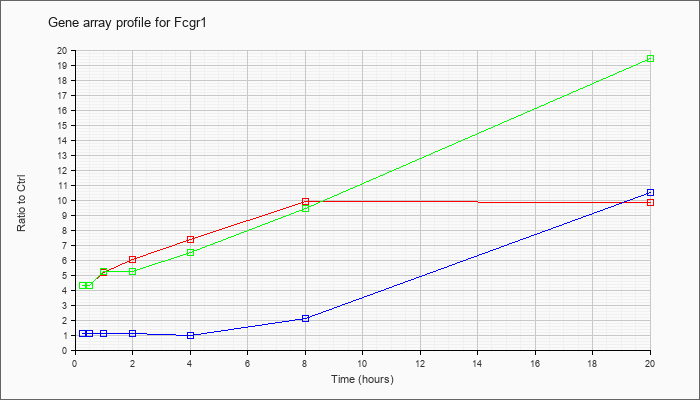 |
AF143181 |
strain AB/H (Biozzi) high affinity immunoglobulin gamma Fc receptor I (Fcgr1) mRNA, Fcgr1-d allele, complete cds. [AF143181] |
KLA | 4.27 |
4.21 |
5.59 |
6.14 |
8.13 |
9.76 |
10.98 |
| ATP | 1.23 |
1.12 |
1.08 |
1.36 |
1.09 |
2.09 |
11.81 |
| KLA/ATP | 4.56 |
4.93 |
4.95 |
5.88 |
6.66 |
9.64 |
23.09 |
|
| Fcgr1 |  |
NM_010186 |
Fc receptor, IgG, high affinity I (Fcgr1), mRNA [NM_010186] |
KLA | 4.31 |
4.37 |
4.70 |
5.97 |
6.59 |
10.06 |
8.67 |
| ATP | 1.01 |
.88 |
1.16 |
.89 |
.90 |
2.15 |
9.23 |
| KLA/ATP | 4.01 |
4.10 |
5.52 |
4.54 |
6.31 |
9.24 |
15.83 |
|
| Fcgr2b | 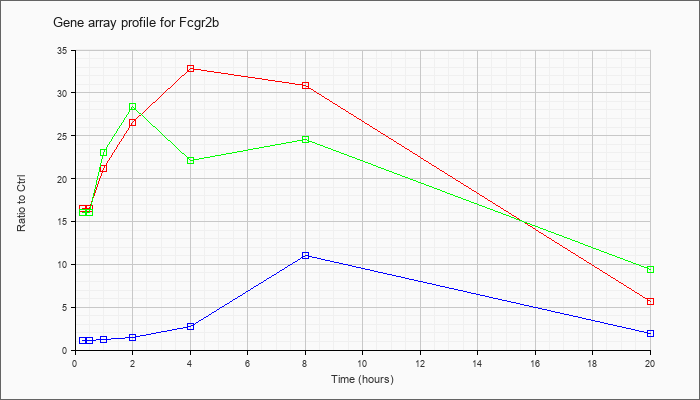 |
NM_001077189 |
Fc receptor, IgG, low affinity IIb (Fcgr2b), transcript variant 1, mRNA [NM_001077189] |
KLA | 16.93 |
17.57 |
21.75 |
27.43 |
34.13 |
31.88 |
5.70 |
| ATP | 1.11 |
1.09 |
1.17 |
1.46 |
2.81 |
11.26 |
1.92 |
| KLA/ATP | 16.51 |
17.91 |
23.67 |
29.22 |
22.67 |
25.33 |
9.54 |
|
| Fcgr2b |  |
NM_010187 |
Fc receptor, IgG, low affinity IIb (Fcgr2b), transcript variant 2, mRNA [NM_010187] |
KLA | 11.41 |
11.67 |
14.06 |
15.47 |
17.28 |
18.63 |
4.69 |
| ATP | 1.05 |
1.06 |
1.18 |
1.57 |
2.27 |
8.97 |
2.10 |
| KLA/ATP | 10.98 |
12.01 |
15.03 |
18.38 |
15.15 |
16.00 |
8.33 |
|
| Gab2 | 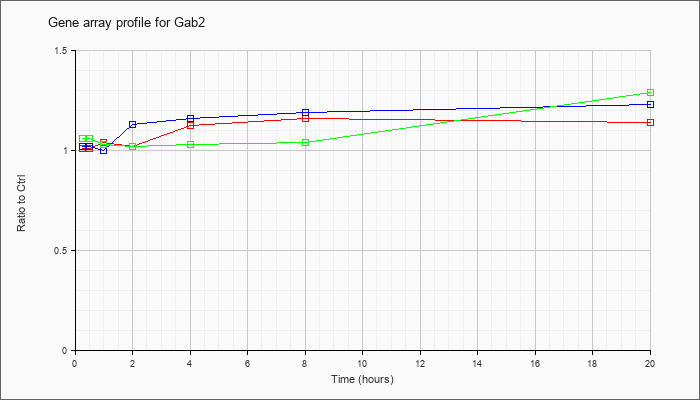 |
NM_010248 |
growth factor receptor bound protein 2-associated protein 2 (Gab2), mRNA [NM_010248] |
KLA | 1.01 |
1.04 |
1.04 |
1.02 |
1.12 |
1.16 |
1.14 |
| ATP | 1.02 |
1.06 |
1.00 |
1.13 |
1.16 |
1.19 |
1.23 |
| KLA/ATP | 1.06 |
1.05 |
1.03 |
1.02 |
1.03 |
1.04 |
1.29 |
|
| Gsn | 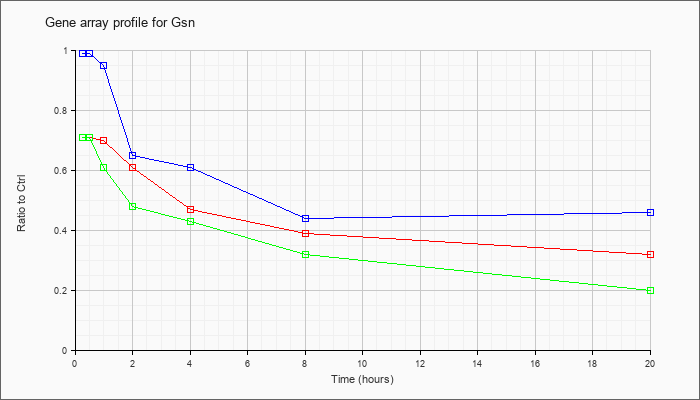 |
NM_146120 |
gelsolin (Gsn), mRNA [NM_146120] |
KLA | .71 |
.69 |
.70 |
.61 |
.47 |
.39 |
.32 |
| ATP | .99 |
.86 |
.95 |
.65 |
.61 |
.44 |
.46 |
| KLA/ATP | .71 |
.80 |
.61 |
.48 |
.43 |
.32 |
.20 |
|
| Hck | 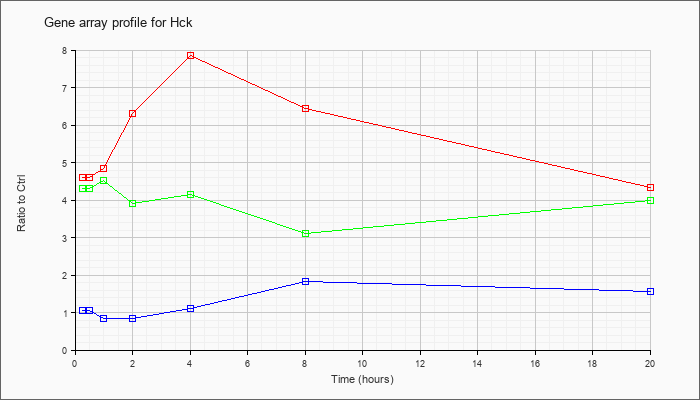 |
NM_010407 |
hemopoietic cell kinase (Hck), mRNA [NM_010407] |
KLA | 4.59 |
4.25 |
4.84 |
6.31 |
7.85 |
6.43 |
4.34 |
| ATP | 1.05 |
1.02 |
.84 |
.84 |
1.12 |
1.82 |
1.55 |
| KLA/ATP | 4.31 |
4.18 |
4.53 |
3.92 |
4.14 |
3.10 |
4.00 |
|
Igh-VJ55
8 |  |
AY547436 |
immunoglobulin heavy chain VDJ region mRNA, partial cds [AY547436] |
KLA | 1.06 |
1.05 |
1.01 |
.93 |
1.03 |
.96 |
.93 |
| ATP | .99 |
1.00 |
1.06 |
.95 |
.96 |
1.06 |
1.05 |
| KLA/ATP | .98 |
.94 |
1.07 |
.95 |
.97 |
.99 |
1.02 |
|
Igh-VJ55
8 |  |
CA578712 |
gb|K0727F04-5N NIA Mouse Hematopoietic Stem Cell (Lin- [CA578712] |
KLA | .99 |
.94 |
1.01 |
1.10 |
.93 |
.97 |
.92 |
| ATP | .93 |
1.04 |
1.01 |
.98 |
1.05 |
.96 |
.95 |
| KLA/ATP | .99 |
.95 |
.95 |
.98 |
1.02 |
.98 |
.95 |
|
Igh-VJ55
8 |  |
ENSMUST00000103412 |
ens|Immunoglobulin heavy chain C gene segment [Source:IMGT/GENE_DB;Acc:IGHA] [ENSMUST00000103412] |
KLA | 1.01 |
.97 |
1.20 |
1.01 |
.98 |
1.44 |
1.24 |
| ATP | .99 |
.98 |
1.07 |
1.03 |
1.00 |
1.01 |
1.21 |
| KLA/ATP | .99 |
1.02 |
.96 |
1.03 |
.97 |
1.13 |
1.30 |
|
Igh-VJ55
8 |  |
ENSMUST00000103413 |
ens|Immunoglobulin heavy chain C gene segment [Source:IMGT/GENE_DB;Acc:IGHE] [ENSMUST00000103413] |
KLA | 1.00 |
.99 |
1.06 |
1.06 |
.99 |
1.03 |
1.02 |
| ATP | 1.08 |
1.06 |
1.06 |
1.06 |
1.02 |
1.06 |
.95 |
| KLA/ATP | 1.00 |
1.04 |
1.09 |
1.02 |
1.06 |
1.02 |
.95 |
|
Igh-VJ55
8 |  |
U39781 |
J558+ IgM heavy chain mRNA, hybridoma clone ME2B7, partial cds. [U39781] |
KLA | .93 |
.89 |
.98 |
.97 |
1.03 |
1.00 |
.94 |
| ATP | 1.04 |
1.02 |
1.08 |
.94 |
.93 |
.90 |
.79 |
| KLA/ATP | .88 |
.89 |
.97 |
.93 |
.88 |
.92 |
.82 |
|
| Inpp5d | 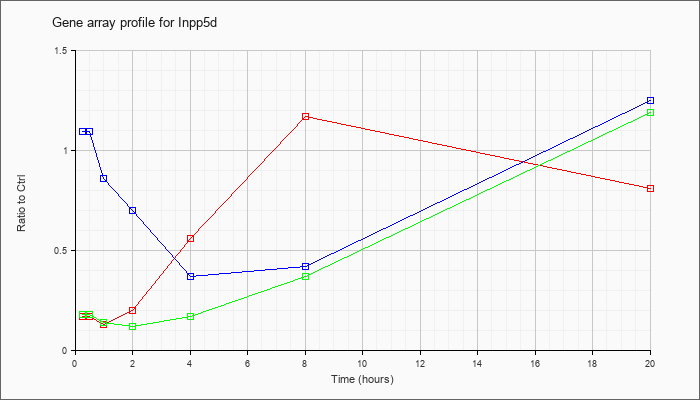 |
NM_010566 |
inositol polyphosphate-5-phosphatase D (Inpp5d), transcript variant 1, mRNA [NM_010566] |
KLA | .17 |
.15 |
.13 |
.20 |
.56 |
1.17 |
.81 |
| ATP | 1.09 |
1.05 |
.86 |
.70 |
.37 |
.42 |
1.25 |
| KLA/ATP | .18 |
.15 |
.14 |
.12 |
.17 |
.37 |
1.19 |
|
| Inppl1 | 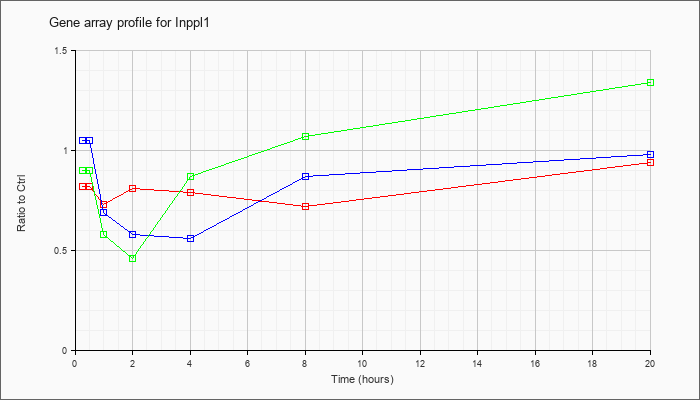 |
NM_010567 |
inositol polyphosphate phosphatase-like 1 (Inppl1), transcript variant 1, mRNA [NM_010567] |
KLA | .82 |
.80 |
.73 |
.81 |
.79 |
.72 |
.94 |
| ATP | 1.05 |
1.04 |
.69 |
.58 |
.56 |
.87 |
.98 |
| KLA/ATP | .90 |
.83 |
.58 |
.46 |
.87 |
1.07 |
1.34 |
|
| Lat | 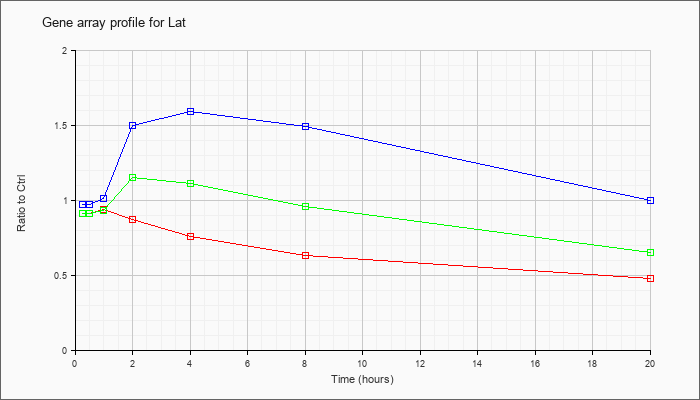 |
NM_010689 |
linker for activation of T cells (Lat), mRNA [NM_010689] |
KLA | .91 |
.95 |
.94 |
.87 |
.76 |
.63 |
.48 |
| ATP | .97 |
.99 |
1.01 |
1.50 |
1.59 |
1.49 |
1.00 |
| KLA/ATP | .91 |
1.04 |
.93 |
1.15 |
1.11 |
.96 |
.65 |
|
| Limk1 | 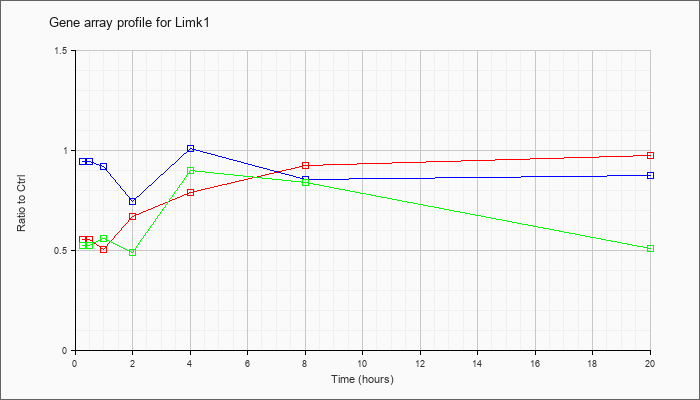 |
NM_010717 |
LIM-domain containing, protein kinase (Limk1), mRNA [NM_010717] |
KLA | .56 |
.57 |
.51 |
.67 |
.79 |
.93 |
.98 |
| ATP | .95 |
.88 |
.92 |
.75 |
1.01 |
.86 |
.88 |
| KLA/ATP | .53 |
.52 |
.56 |
.49 |
.90 |
.84 |
.51 |
|
| Limk2 | 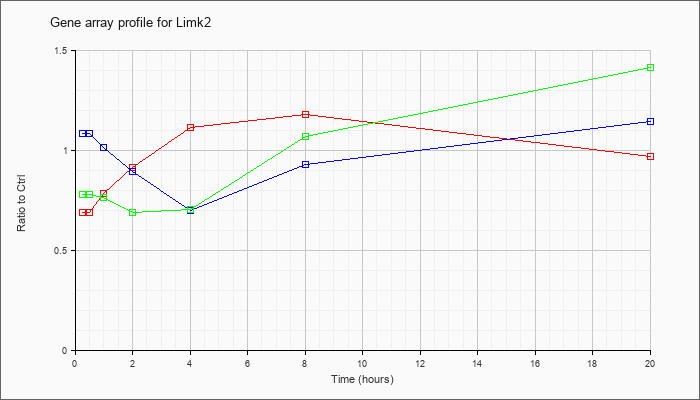 |
NM_010718 |
LIM motif-containing protein kinase 2 (Limk2), transcript variant 1, mRNA [NM_010718] |
KLA | .69 |
.73 |
.79 |
.92 |
1.12 |
1.18 |
.97 |
| ATP | 1.09 |
1.05 |
1.02 |
.90 |
.70 |
.93 |
1.15 |
| KLA/ATP | .78 |
.76 |
.77 |
.69 |
.71 |
1.07 |
1.42 |
|
| Lyn | 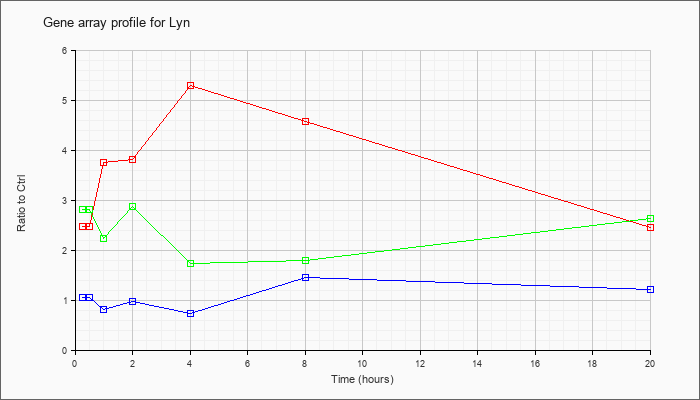 |
NM_010747 |
Yamaguchi sarcoma viral (v-yes-1) oncogene homolog (Lyn), transcript variant 2, mRNA [NM_010747] |
KLA | 2.48 |
2.80 |
3.76 |
3.81 |
5.30 |
4.57 |
2.46 |
| ATP | 1.06 |
1.18 |
.82 |
.97 |
.74 |
1.45 |
1.21 |
| KLA/ATP | 2.82 |
3.16 |
2.24 |
2.87 |
1.73 |
1.80 |
2.63 |
|
| Map2k1 | 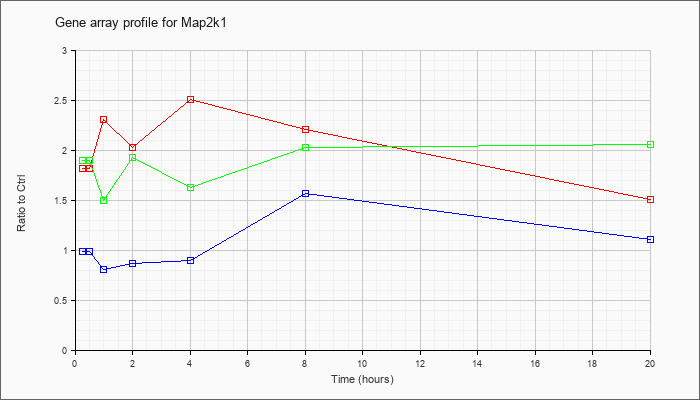 |
NM_008927 |
mitogen-activated protein kinase kinase 1 (Map2k1), mRNA [NM_008927] |
KLA | 1.82 |
2.00 |
2.31 |
2.03 |
2.51 |
2.20 |
1.51 |
| ATP | .99 |
1.14 |
.81 |
.87 |
.90 |
1.57 |
1.11 |
| KLA/ATP | 1.90 |
2.03 |
1.50 |
1.93 |
1.63 |
2.03 |
2.06 |
|
| Mapk1 | 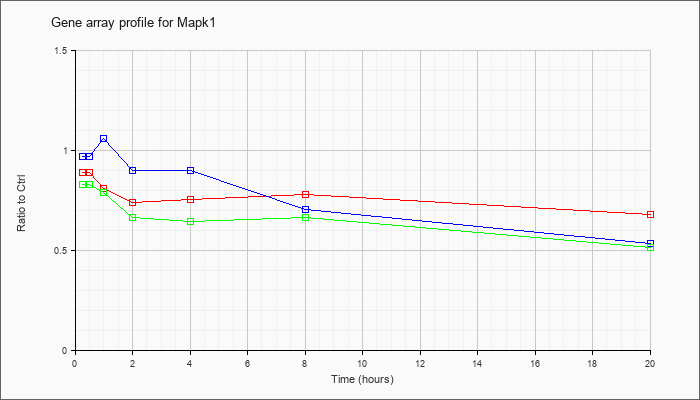 |
NM_001038663 |
mitogen-activated protein kinase 1 (Mapk1), transcript variant 2, mRNA [NM_001038663] |
KLA | .97 |
1.03 |
.93 |
.86 |
.79 |
1.01 |
1.04 |
| ATP | .96 |
1.18 |
1.01 |
1.13 |
.84 |
1.04 |
1.16 |
| KLA/ATP | .99 |
.99 |
.84 |
.87 |
.63 |
.96 |
1.26 |
|
| Mapk1 |  |
NM_011949 |
mitogen-activated protein kinase 1 (Mapk1), transcript variant 1, mRNA [NM_011949] |
KLA | .88 |
.80 |
.80 |
.73 |
.75 |
.76 |
.65 |
| ATP | .97 |
.96 |
1.06 |
.88 |
.90 |
.68 |
.48 |
| KLA/ATP | .82 |
.81 |
.78 |
.65 |
.64 |
.64 |
.45 |
|
| Mapk3 | 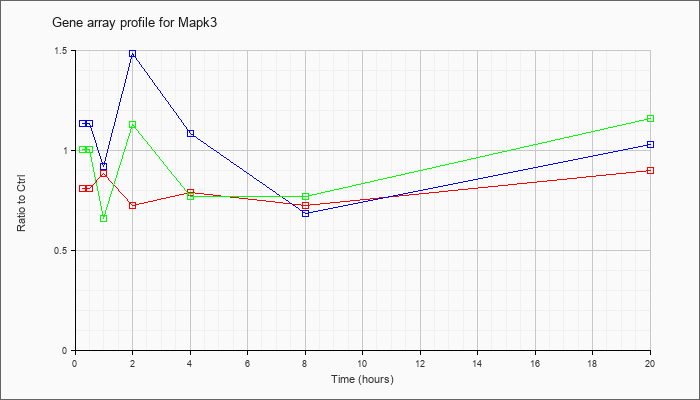 |
NM_011952 |
mitogen-activated protein kinase 3 (Mapk3), mRNA [NM_011952] |
KLA | .81 |
.85 |
.88 |
.72 |
.79 |
.73 |
.90 |
| ATP | 1.13 |
1.30 |
.92 |
1.48 |
1.08 |
.68 |
1.03 |
| KLA/ATP | 1.01 |
1.01 |
.66 |
1.13 |
.77 |
.77 |
1.16 |
|
| Marcks | 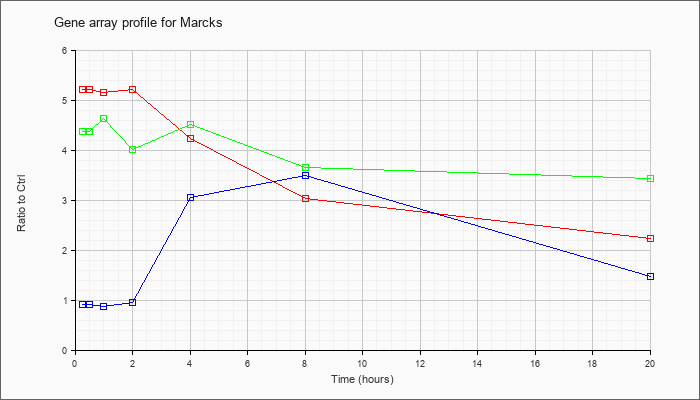 |
NM_008538 |
myristoylated alanine rich protein kinase C substrate (Marcks), mRNA [NM_008538] |
KLA | 5.22 |
4.64 |
5.16 |
5.22 |
4.23 |
3.04 |
2.24 |
| ATP | .92 |
.90 |
.87 |
.95 |
3.05 |
3.49 |
1.47 |
| KLA/ATP | 4.38 |
4.30 |
4.63 |
4.01 |
4.51 |
3.65 |
3.44 |
|
Marcksl1
| 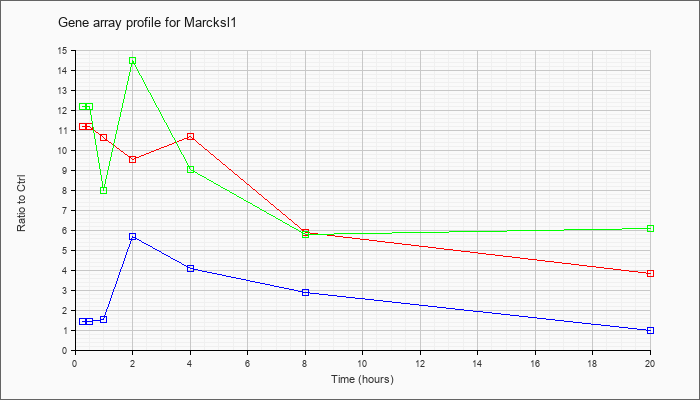 |
NM_010807 |
MARCKS-like 1 (Marcksl1), mRNA [NM_010807] |
KLA | 11.17 |
9.95 |
10.63 |
9.53 |
10.67 |
5.90 |
3.84 |
| ATP | 1.42 |
1.91 |
1.55 |
5.70 |
4.10 |
2.89 |
.97 |
| KLA/ATP | 12.20 |
13.11 |
7.97 |
14.47 |
9.03 |
5.78 |
6.06 |
|
Mm.21312
8 | 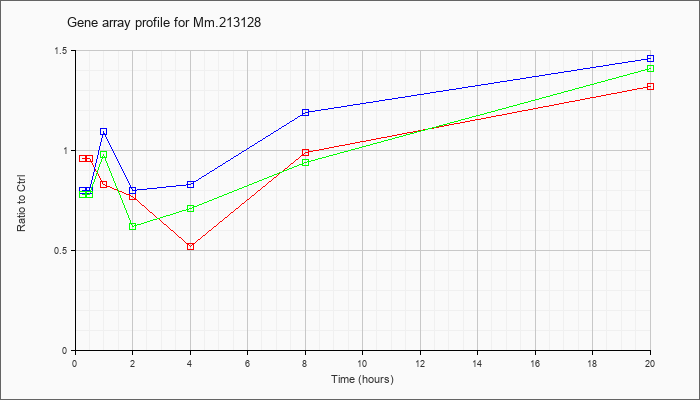 |
141801344 |
Unknown |
KLA | .96 |
.95 |
.83 |
.77 |
.52 |
.99 |
1.32 |
| ATP | .80 |
.82 |
1.09 |
.80 |
.83 |
1.19 |
1.46 |
| KLA/ATP | .78 |
.92 |
.98 |
.62 |
.71 |
.94 |
1.41 |
|
Mm.25381
9 | 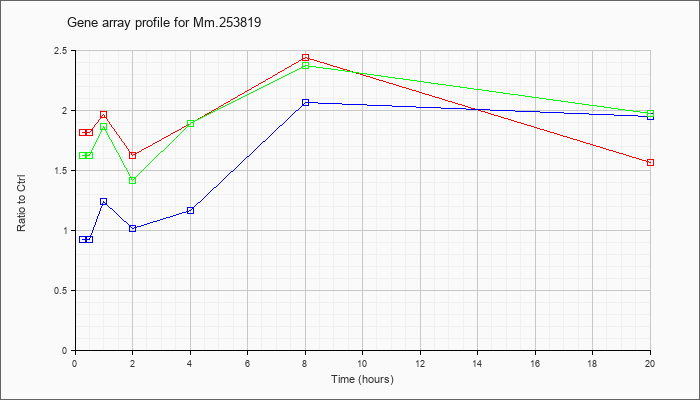 |
118130676 |
Unknown |
KLA | 1.81 |
1.88 |
1.96 |
1.62 |
1.89 |
2.44 |
1.56 |
| ATP | .92 |
.91 |
1.24 |
1.01 |
1.16 |
2.06 |
1.95 |
| KLA/ATP | 1.62 |
1.86 |
1.86 |
1.41 |
1.89 |
2.37 |
1.97 |
|
Mm.27682
6 | 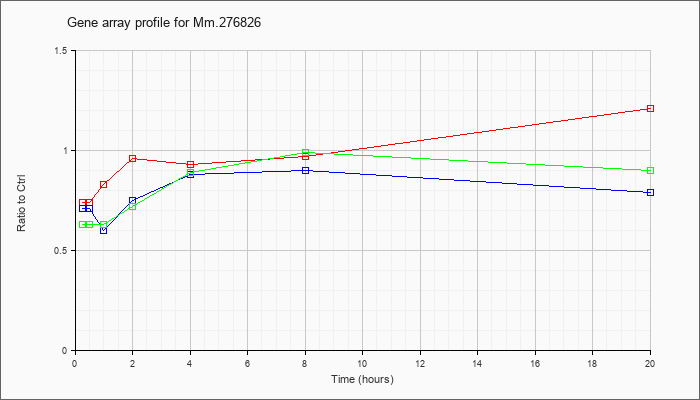 |
126513132 |
Unknown |
KLA | .74 |
.69 |
.83 |
.96 |
.93 |
.97 |
1.21 |
| ATP | .71 |
.66 |
.60 |
.75 |
.88 |
.90 |
.79 |
| KLA/ATP | .63 |
.57 |
.63 |
.72 |
.89 |
.99 |
.90 |
|
Mm.28873
| 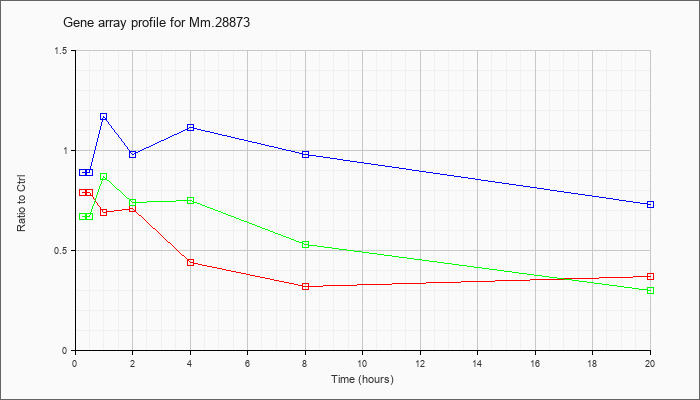 |
110431340 |
Unknown |
KLA | .79 |
.72 |
.69 |
.71 |
.44 |
.32 |
.37 |
| ATP | .89 |
.88 |
1.17 |
.98 |
1.11 |
.98 |
.73 |
| KLA/ATP | .67 |
.72 |
.87 |
.74 |
.75 |
.53 |
.30 |
|
Mm.35894
6 | 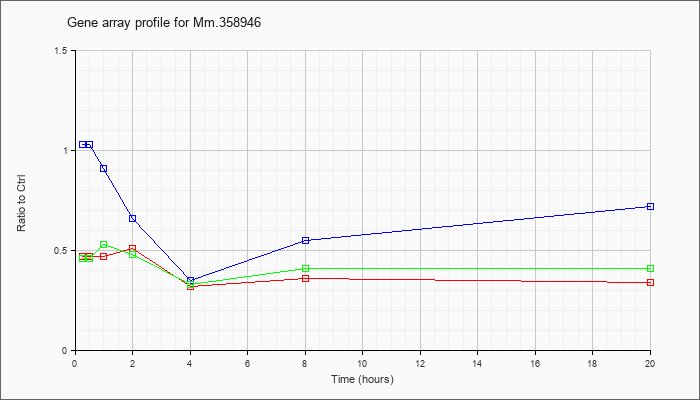 |
147907211 |
Unknown |
KLA | .47 |
.50 |
.47 |
.51 |
.32 |
.36 |
.34 |
| ATP | 1.03 |
.98 |
.91 |
.66 |
.35 |
.55 |
.72 |
| KLA/ATP | .46 |
.47 |
.53 |
.48 |
.33 |
.41 |
.41 |
|
Mm.37161
0 | 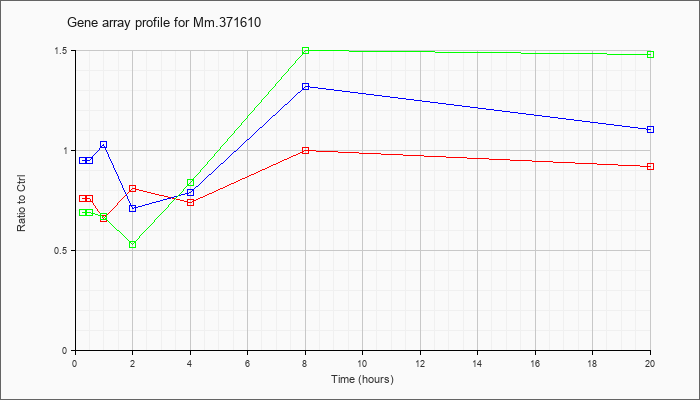 |
118130576 |
Unknown |
KLA | .76 |
.74 |
.66 |
.81 |
.74 |
1.00 |
.92 |
| ATP | .95 |
.85 |
1.03 |
.71 |
.79 |
1.32 |
1.10 |
| KLA/ATP | .69 |
.70 |
.67 |
.53 |
.84 |
1.50 |
1.48 |
|
Mm.42529
6 | 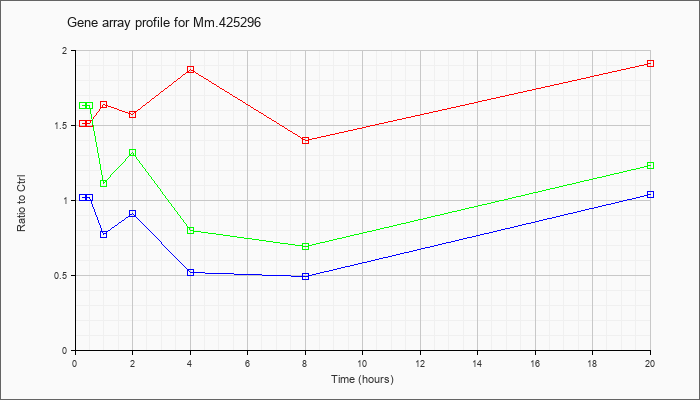 |
170172552 |
Unknown |
KLA | 1.51 |
1.59 |
1.64 |
1.57 |
1.87 |
1.40 |
1.91 |
| ATP | 1.02 |
1.04 |
.77 |
.91 |
.52 |
.49 |
1.04 |
| KLA/ATP | 1.63 |
1.53 |
1.11 |
1.32 |
.80 |
.69 |
1.23 |
|
Mm.44463
|  |
118129851 |
Unknown |
KLA | 1.01 |
.98 |
.97 |
1.02 |
.93 |
1.06 |
1.05 |
| ATP | .97 |
1.01 |
1.04 |
1.06 |
1.02 |
1.08 |
.97 |
| KLA/ATP | 1.02 |
1.06 |
.95 |
1.04 |
1.09 |
1.14 |
1.03 |
|
| Myo10 |  |
AK089482 |
B6-derived CD11 +ve dendritic cells cDNA, RIKEN full-length enriched library, clone:F730038D06 product:hypothetical protein, full insert sequence. [AK089482] |
KLA | 1.21 |
1.09 |
1.19 |
1.04 |
1.11 |
1.26 |
1.34 |
| ATP | 1.01 |
.96 |
1.26 |
1.32 |
1.22 |
1.20 |
1.10 |
| KLA/ATP | 1.17 |
1.15 |
1.27 |
1.21 |
1.06 |
1.14 |
1.37 |
|
| Myo10 |  |
NM_019472 |
myosin X (Myo10), mRNA [NM_019472] |
KLA | 3.54 |
3.54 |
2.96 |
2.33 |
1.61 |
1.36 |
1.34 |
| ATP | 1.04 |
1.01 |
.60 |
.79 |
.98 |
1.03 |
1.17 |
| KLA/ATP | 3.88 |
3.56 |
1.75 |
1.75 |
.97 |
.86 |
2.22 |
|
| Ncf1 |  |
NM_010876 |
neutrophil cytosolic factor 1 (Ncf1), mRNA [NM_010876] |
KLA | 1.62 |
1.71 |
1.81 |
1.70 |
1.99 |
1.64 |
2.07 |
| ATP | 1.02 |
1.06 |
.78 |
1.02 |
.64 |
.56 |
1.08 |
| KLA/ATP | 1.87 |
1.81 |
1.22 |
1.73 |
1.00 |
.80 |
1.49 |
|
| Pak1 |  |
NM_011035 |
p21 (CDKN1A)-activated kinase 1 (Pak1), mRNA [NM_011035] |
KLA | .72 |
.77 |
.79 |
.71 |
.67 |
.73 |
.95 |
| ATP | 1.10 |
1.07 |
.99 |
1.15 |
1.27 |
.62 |
.51 |
| KLA/ATP | .85 |
.83 |
.73 |
.76 |
.76 |
.55 |
.33 |
|
| Pik3ca | 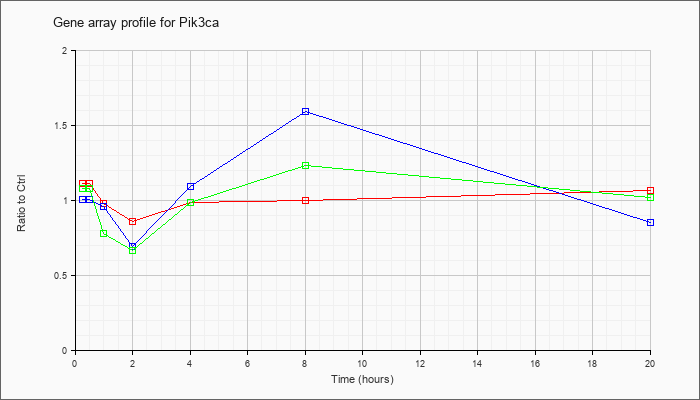 |
ENSMUST00000108243 |
ens|Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). [Source:Uniprot/SWISSPROT;Acc:P42337] [ENSMUST00000108243] |
KLA | 1.07 |
1.01 |
.91 |
.90 |
.98 |
.96 |
.86 |
| ATP | 1.03 |
.87 |
1.12 |
.63 |
1.22 |
1.86 |
.60 |
| KLA/ATP | 1.05 |
.82 |
.91 |
.61 |
1.17 |
1.26 |
.63 |
|
| Pik3ca |  |
NM_008839 |
phosphatidylinositol 3-kinase, catalytic, alpha polypeptide (Pik3ca), mRNA [NM_008839] |
KLA | 1.15 |
1.11 |
1.05 |
.82 |
.99 |
1.03 |
1.27 |
| ATP | .98 |
1.00 |
.79 |
.75 |
.96 |
1.32 |
1.10 |
| KLA/ATP | 1.10 |
.98 |
.64 |
.72 |
.80 |
1.20 |
1.41 |
|
| Pik3cb | 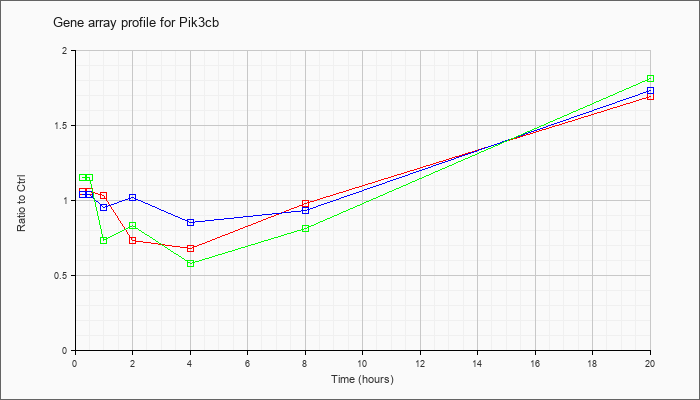 |
NM_029094 |
phosphatidylinositol 3-kinase, catalytic, beta polypeptide (Pik3cb), mRNA [NM_029094] |
KLA | 1.06 |
1.06 |
1.03 |
.73 |
.68 |
.98 |
1.69 |
| ATP | 1.04 |
1.21 |
.95 |
1.02 |
.85 |
.93 |
1.73 |
| KLA/ATP | 1.15 |
1.27 |
.73 |
.83 |
.58 |
.81 |
1.81 |
|
| Pik3cd | 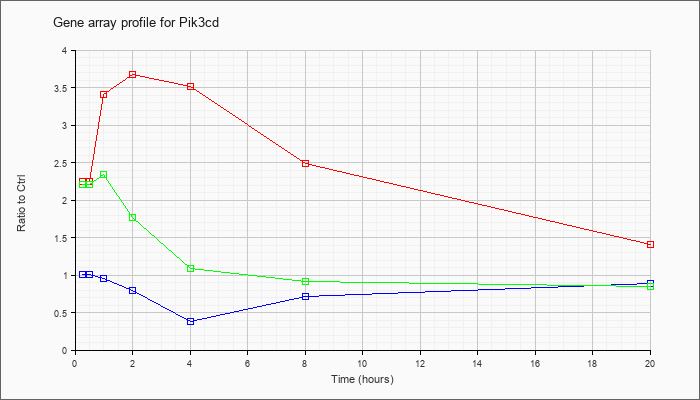 |
NM_008840 |
phosphatidylinositol 3-kinase catalytic delta polypeptide (Pik3cd), transcript variant 1, mRNA [NM_008840] |
KLA | 2.28 |
2.42 |
3.10 |
4.17 |
3.35 |
2.31 |
1.34 |
| ATP | 1.02 |
.91 |
.99 |
.63 |
.30 |
.67 |
.80 |
| KLA/ATP | 2.14 |
2.23 |
2.70 |
1.51 |
1.17 |
.85 |
.72 |
|
| Pik3cd |  |
U86587 |
phosphatidylinositol 3-kinase catalytic subunit p110 delta mRNA, complete cds. [U86587] |
KLA | 2.21 |
2.24 |
3.71 |
3.17 |
3.67 |
2.66 |
1.48 |
| ATP | 1.00 |
1.03 |
.93 |
.96 |
.46 |
.75 |
.97 |
| KLA/ATP | 2.27 |
2.32 |
1.97 |
2.02 |
1.01 |
.99 |
.98 |
|
| Pik3cg | 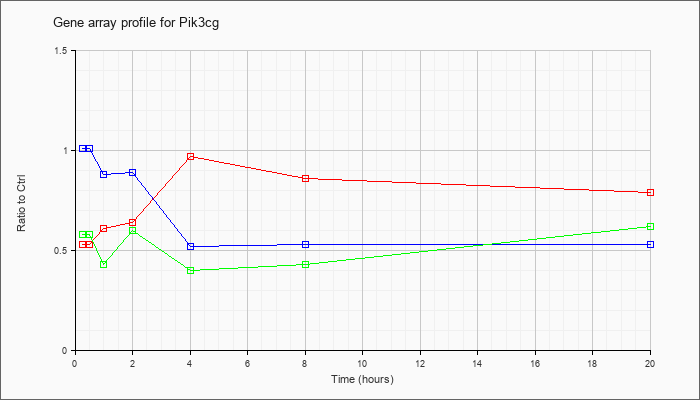 |
NM_020272 |
phosphoinositide-3-kinase, catalytic, gamma polypeptide (Pik3cg), mRNA [NM_020272] |
KLA | .53 |
.53 |
.61 |
.64 |
.97 |
.86 |
.79 |
| ATP | 1.01 |
1.13 |
.88 |
.89 |
.52 |
.53 |
.53 |
| KLA/ATP | .58 |
.53 |
.43 |
.60 |
.40 |
.43 |
.62 |
|
| Pik3r1 | 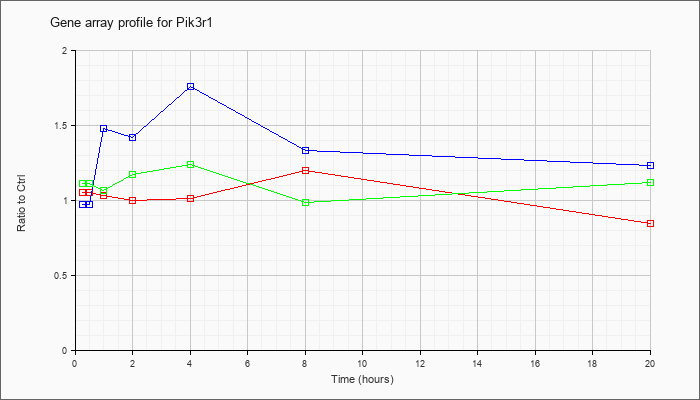 |
NM_001077495 |
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) (Pik3r1), transcript variant 2, mRNA [NM_001077495] |
KLA | 1.05 |
1.07 |
1.03 |
1.00 |
1.01 |
1.20 |
.85 |
| ATP | .97 |
1.02 |
1.48 |
1.42 |
1.76 |
1.33 |
1.23 |
| KLA/ATP | 1.11 |
1.12 |
1.06 |
1.17 |
1.24 |
.99 |
1.12 |
|
| Pik3r2 | 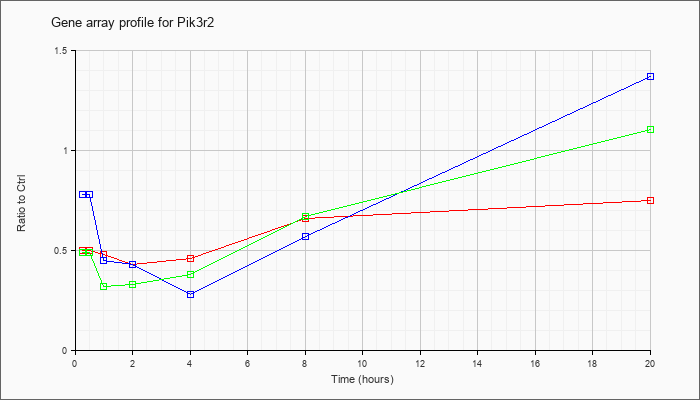 |
NM_008841 |
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 2 (p85 beta) (Pik3r2), mRNA [NM_008841] |
KLA | .50 |
.50 |
.48 |
.43 |
.46 |
.66 |
.75 |
| ATP | .78 |
.68 |
.45 |
.43 |
.28 |
.57 |
1.37 |
| KLA/ATP | .49 |
.38 |
.32 |
.33 |
.38 |
.67 |
1.10 |
|
| Pik3r3 | 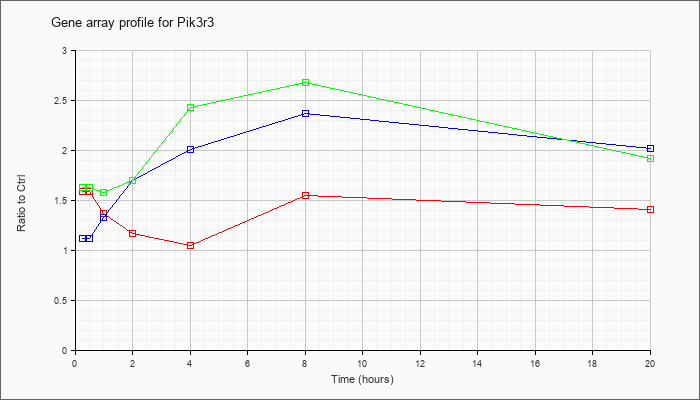 |
NM_181585 |
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) (Pik3r3), mRNA [NM_181585] |
KLA | 1.59 |
1.49 |
1.37 |
1.17 |
1.05 |
1.55 |
1.41 |
| ATP | 1.12 |
1.00 |
1.33 |
1.70 |
2.01 |
2.37 |
2.02 |
| KLA/ATP | 1.63 |
1.54 |
1.58 |
1.70 |
2.43 |
2.68 |
1.92 |
|
| Pik3r5 | 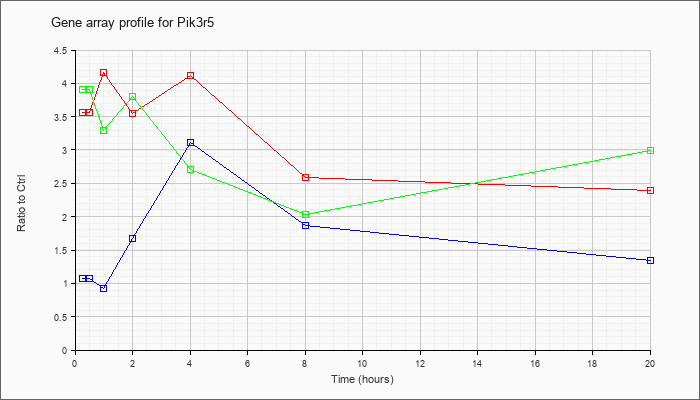 |
NM_177320 |
phosphoinositide-3-kinase, regulatory subunit 5, p101 (Pik3r5), mRNA [NM_177320] |
KLA | 3.57 |
3.58 |
4.16 |
3.54 |
4.11 |
2.59 |
2.39 |
| ATP | 1.08 |
1.08 |
.93 |
1.67 |
3.11 |
1.88 |
1.34 |
| KLA/ATP | 3.91 |
3.70 |
3.30 |
3.80 |
2.70 |
2.04 |
3.00 |
|
| Pip5k1a | 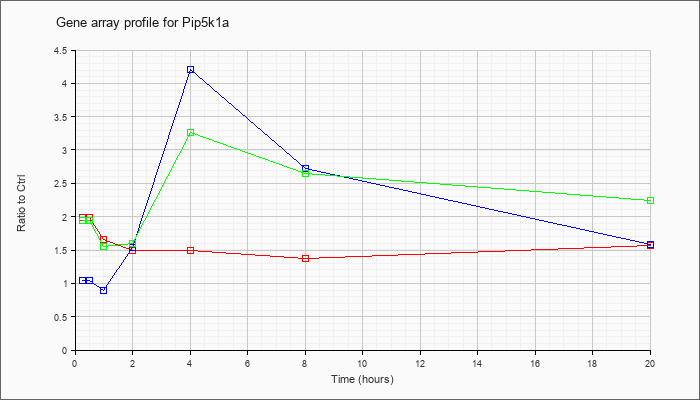 |
NM_008847 |
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha (Pip5k1a), mRNA [NM_008847] |
KLA | 1.99 |
1.84 |
1.66 |
1.49 |
1.49 |
1.37 |
1.57 |
| ATP | 1.05 |
1.06 |
.89 |
1.54 |
4.20 |
2.72 |
1.58 |
| KLA/ATP | 1.95 |
1.90 |
1.55 |
1.60 |
3.26 |
2.64 |
2.25 |
|
| Pip5k1b | 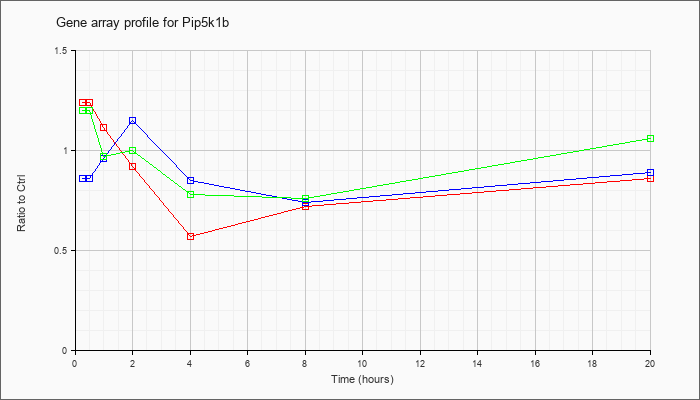 |
NM_008846 |
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta (Pip5k1b), mRNA [NM_008846] |
KLA | 1.24 |
1.35 |
1.11 |
.92 |
.57 |
.72 |
.86 |
| ATP | .86 |
1.00 |
.96 |
1.15 |
.85 |
.74 |
.89 |
| KLA/ATP | 1.20 |
1.22 |
.97 |
1.00 |
.78 |
.76 |
1.06 |
|
| Pip5k1c | 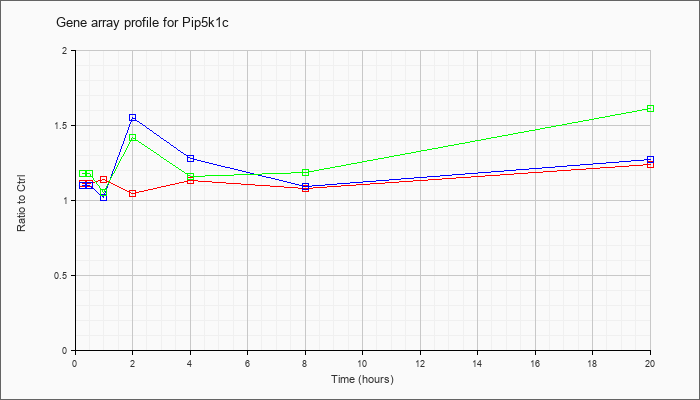 |
BC094665 |
phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma, mRNA (cDNA clone MGC:102155 IMAGE:30356181), complete cds [BC094665] |
KLA | .82 |
.90 |
.90 |
.70 |
.96 |
.86 |
1.21 |
| ATP | 1.19 |
1.45 |
.92 |
1.64 |
1.17 |
.83 |
1.33 |
| KLA/ATP | .97 |
1.21 |
.66 |
1.20 |
.91 |
.92 |
1.69 |
|
| Pip5k1c |  |
NM_008844 |
phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma (Pip5k1c), mRNA [NM_008844] |
KLA | 1.40 |
1.36 |
1.38 |
1.39 |
1.30 |
1.30 |
1.27 |
| ATP | 1.01 |
1.16 |
1.12 |
1.46 |
1.38 |
1.35 |
1.21 |
| KLA/ATP | 1.39 |
1.44 |
1.44 |
1.64 |
1.41 |
1.45 |
1.53 |
|
| Pla2g4a | 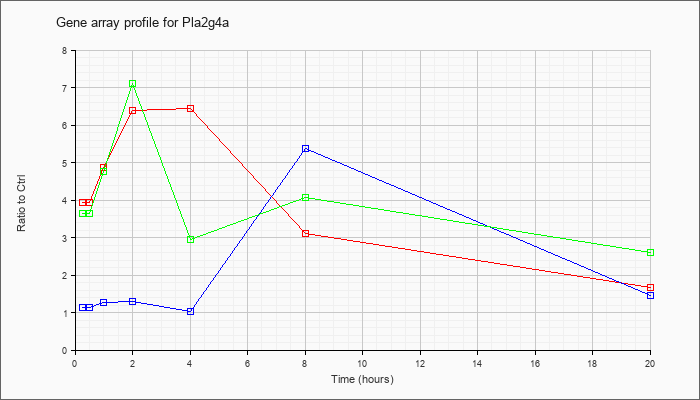 |
NM_008869 |
phospholipase A2, group IVA (cytosolic, calcium-dependent) (Pla2g4a), mRNA [NM_008869] |
KLA | 3.93 |
4.49 |
4.86 |
6.40 |
6.44 |
3.10 |
1.68 |
| ATP | 1.13 |
1.36 |
1.27 |
1.30 |
1.02 |
5.37 |
1.47 |
| KLA/ATP | 3.64 |
4.45 |
4.76 |
7.11 |
2.96 |
4.08 |
2.60 |
|
| Pla2g4e | 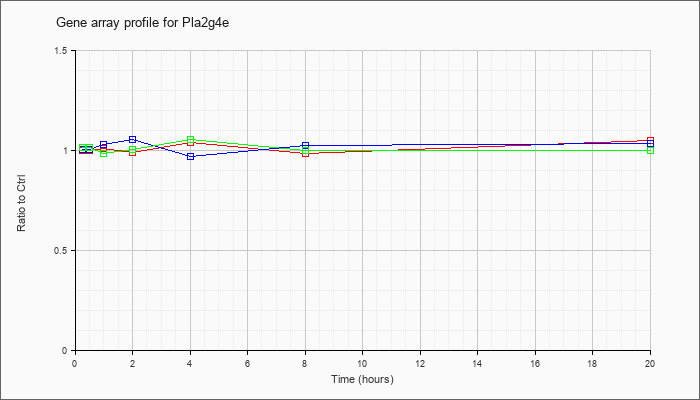 |
AK049063 |
0 day neonate cerebellum cDNA, RIKEN full-length enriched library, clone:C230096D22 product:hypothetical Lysophospholipase catalytic domain containing protein, full insert sequence. [AK049063] |
KLA | 1.03 |
1.09 |
1.01 |
1.01 |
1.06 |
.94 |
1.05 |
| ATP | 1.03 |
1.00 |
1.06 |
1.01 |
.96 |
1.05 |
1.05 |
| KLA/ATP | .98 |
1.00 |
1.01 |
.98 |
1.03 |
.95 |
1.01 |
|
| Pla2g4e |  |
NM_177845 |
phospholipase A2, group IVE (Pla2g4e), mRNA [NM_177845] |
KLA | .97 |
.95 |
1.01 |
.97 |
1.02 |
1.03 |
1.05 |
| ATP | .98 |
.99 |
1.00 |
1.10 |
.98 |
1.00 |
1.02 |
| KLA/ATP | 1.05 |
.93 |
.96 |
1.03 |
1.08 |
1.05 |
.99 |
|
| Pla2g6 | 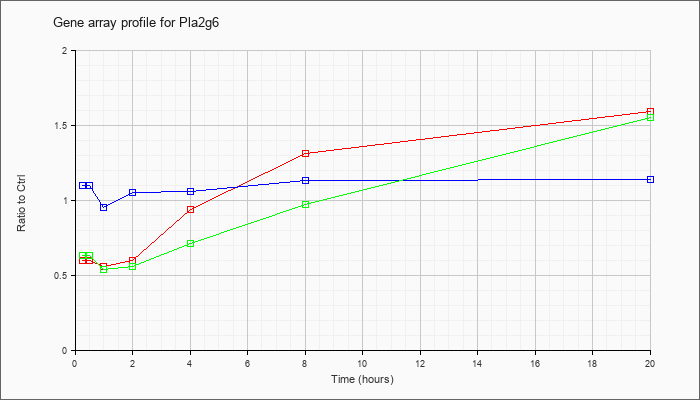 |
NM_016915 |
phospholipase A2, group VI (Pla2g6), mRNA [NM_016915] |
KLA | .60 |
.59 |
.56 |
.60 |
.94 |
1.31 |
1.59 |
| ATP | 1.10 |
1.14 |
.95 |
1.05 |
1.06 |
1.13 |
1.14 |
| KLA/ATP | .63 |
.56 |
.54 |
.56 |
.71 |
.97 |
1.55 |
|
| Plcg2 | 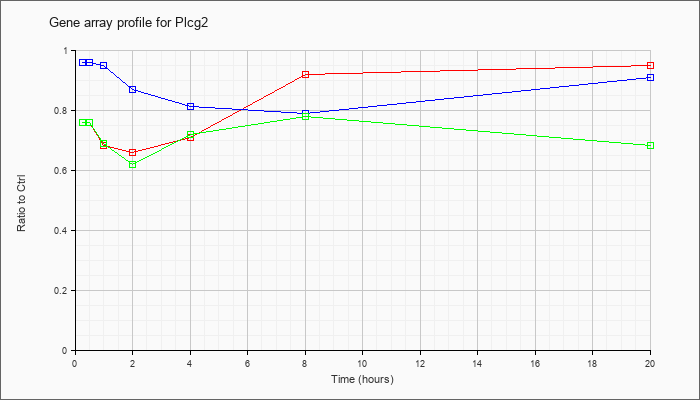 |
NM_172285 |
phospholipase C, gamma 2 (Plcg2), mRNA [NM_172285] |
KLA | .76 |
.74 |
.68 |
.66 |
.71 |
.92 |
.95 |
| ATP | .96 |
.90 |
.95 |
.87 |
.81 |
.79 |
.91 |
| KLA/ATP | .76 |
.71 |
.69 |
.62 |
.72 |
.78 |
.68 |
|
| Pld1 | 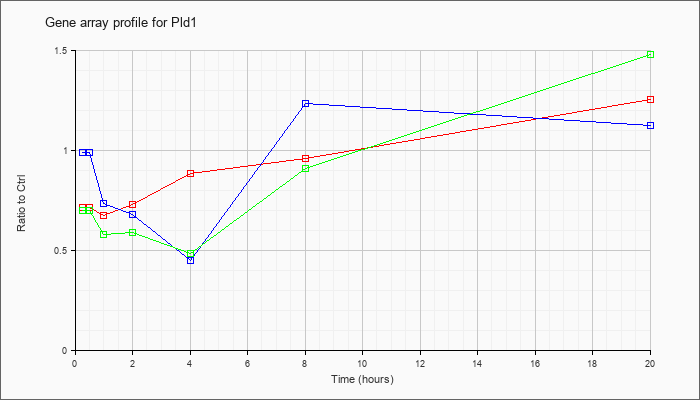 |
NM_008875 |
phospholipase D1 (Pld1), mRNA [NM_008875] |
KLA | .71 |
.73 |
.67 |
.73 |
.88 |
.96 |
1.25 |
| ATP | .99 |
.88 |
.73 |
.68 |
.45 |
1.23 |
1.12 |
| KLA/ATP | .70 |
.67 |
.58 |
.59 |
.48 |
.91 |
1.48 |
|
| Pld2 | 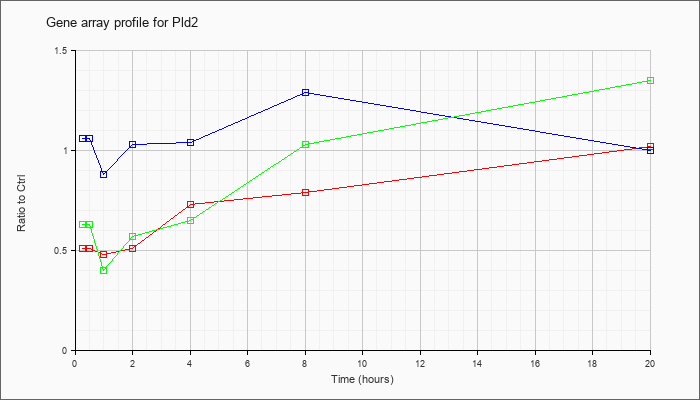 |
NM_008876 |
phospholipase D2 (Pld2), mRNA [NM_008876] |
KLA | .51 |
.54 |
.48 |
.51 |
.73 |
.79 |
1.02 |
| ATP | 1.06 |
1.10 |
.88 |
1.03 |
1.04 |
1.29 |
1.00 |
| KLA/ATP | .63 |
.52 |
.40 |
.57 |
.65 |
1.03 |
1.35 |
|
| Ppap2a | 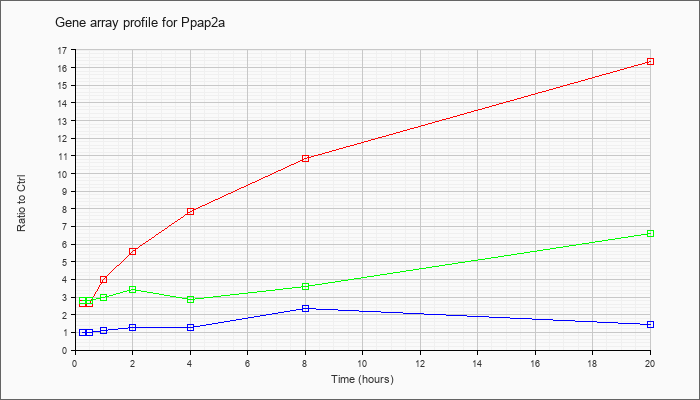 |
NM_008903 |
phosphatidic acid phosphatase 2a (Ppap2a), transcript variant 2, mRNA [NM_008903] |
KLA | 2.62 |
2.86 |
4.01 |
5.61 |
7.84 |
10.88 |
16.35 |
| ATP | 1.01 |
1.02 |
1.09 |
1.28 |
1.27 |
2.38 |
1.44 |
| KLA/ATP | 2.78 |
2.78 |
2.99 |
3.43 |
2.85 |
3.59 |
6.59 |
|
| Ppap2b | 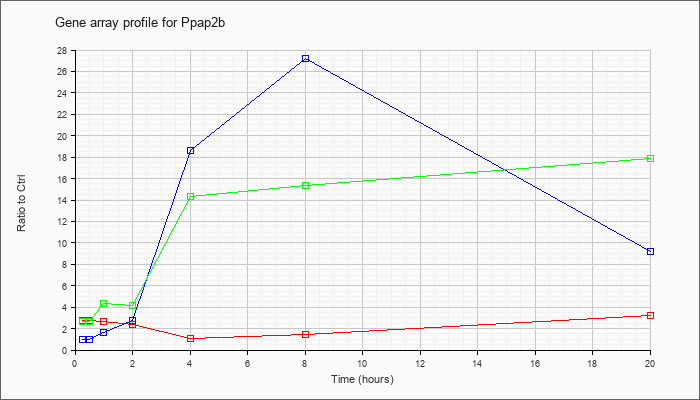 |
NM_080555 |
phosphatidic acid phosphatase type 2B (Ppap2b), mRNA [NM_080555] |
KLA | 2.73 |
2.72 |
2.65 |
2.39 |
1.07 |
1.48 |
3.26 |
| ATP | .99 |
.97 |
1.60 |
2.76 |
18.64 |
27.23 |
9.19 |
| KLA/ATP | 2.69 |
2.80 |
4.36 |
4.20 |
14.33 |
15.32 |
17.91 |
|
| Ppap2c | 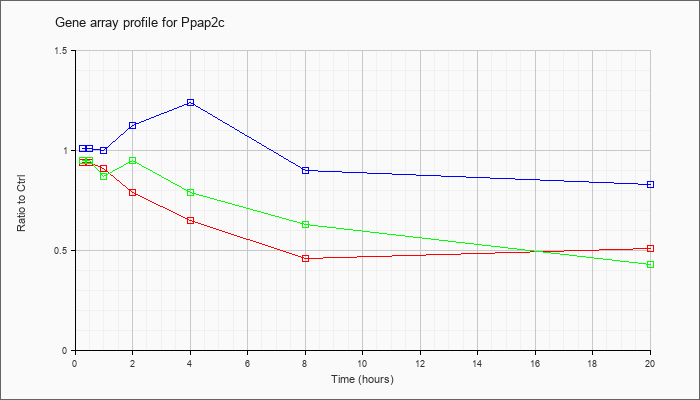 |
NM_015817 |
phosphatidic acid phosphatase type 2c (Ppap2c), mRNA [NM_015817] |
KLA | .94 |
.88 |
.91 |
.79 |
.65 |
.46 |
.51 |
| ATP | 1.01 |
1.03 |
1.00 |
1.12 |
1.24 |
.90 |
.83 |
| KLA/ATP | .95 |
.93 |
.87 |
.95 |
.79 |
.63 |
.43 |
|
| Prkca | 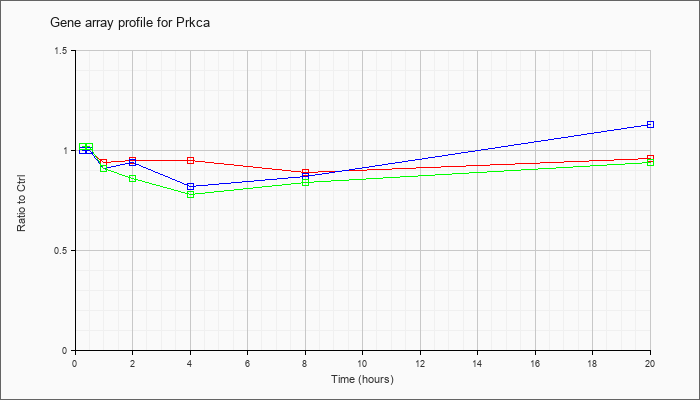 |
NM_011101 |
protein kinase C, alpha (Prkca), mRNA [NM_011101] |
KLA | 1.00 |
.96 |
.94 |
.95 |
.95 |
.89 |
.96 |
| ATP | 1.00 |
1.03 |
.91 |
.94 |
.82 |
.87 |
1.13 |
| KLA/ATP | 1.02 |
.95 |
.91 |
.86 |
.78 |
.84 |
.94 |
|
| Prkcb1 | 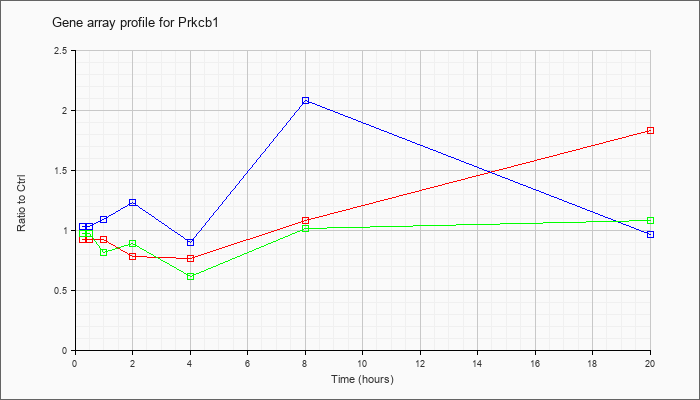 |
NM_008855 |
protein kinase C, beta 1 (Prkcb1), mRNA [NM_008855] |
KLA | .92 |
.95 |
.93 |
.78 |
.77 |
1.08 |
1.83 |
| ATP | 1.03 |
1.12 |
1.09 |
1.23 |
.90 |
2.08 |
.96 |
| KLA/ATP | .98 |
.99 |
.82 |
.89 |
.62 |
1.01 |
1.08 |
|
| Prkcc | 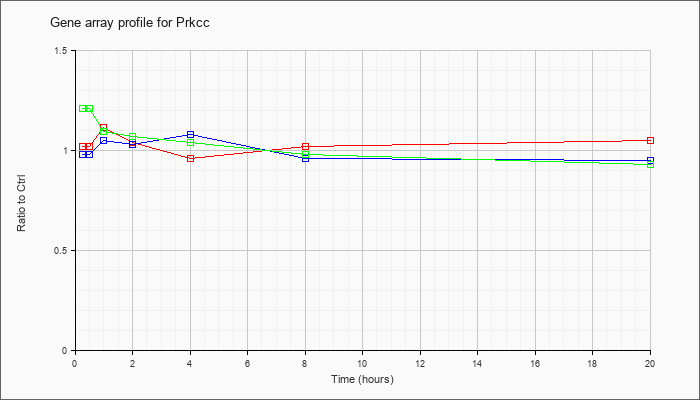 |
NM_011102 |
protein kinase C, gamma (Prkcc), mRNA [NM_011102] |
KLA | 1.02 |
1.04 |
1.11 |
1.04 |
.96 |
1.02 |
1.05 |
| ATP | .98 |
.98 |
1.05 |
1.03 |
1.08 |
.96 |
.95 |
| KLA/ATP | 1.21 |
1.16 |
1.09 |
1.07 |
1.04 |
.98 |
.93 |
|
| Prkcd | 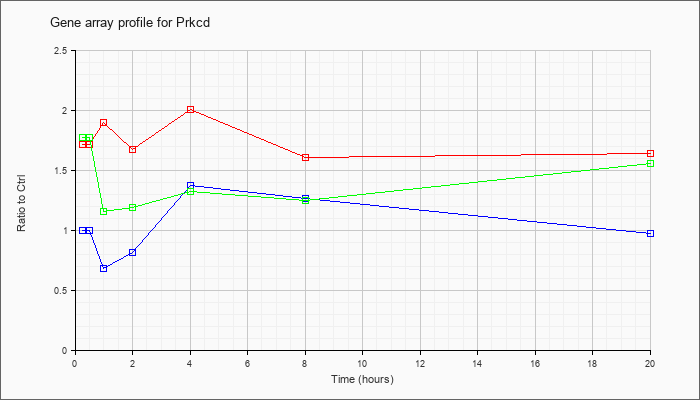 |
NM_011103 |
protein kinase C, delta (Prkcd), mRNA [NM_011103] |
KLA | 1.72 |
1.84 |
1.90 |
1.68 |
2.01 |
1.61 |
1.64 |
| ATP | 1.00 |
1.03 |
.68 |
.81 |
1.38 |
1.26 |
.98 |
| KLA/ATP | 1.77 |
1.79 |
1.16 |
1.19 |
1.33 |
1.25 |
1.56 |
|
| Prkce | 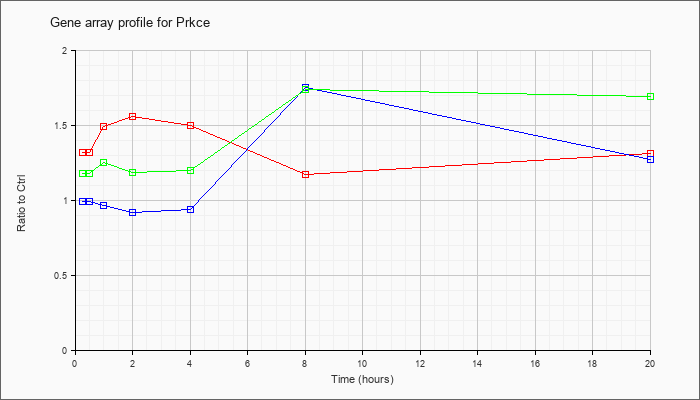 |
AK017901 |
adult male thymus cDNA, RIKEN full-length enriched library, clone:5830406C15 product:unclassifiable, full insert sequence. [AK017901] |
KLA | 1.63 |
1.44 |
1.95 |
2.09 |
1.96 |
1.34 |
1.56 |
| ATP | .99 |
.83 |
.85 |
.78 |
.85 |
2.44 |
1.47 |
| KLA/ATP | 1.36 |
1.45 |
1.44 |
1.23 |
1.30 |
2.39 |
2.29 |
|
| Prkce |  |
AK042994 |
7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730046G04 product:protein kinase C, epsilon, full insert sequence. [AK042994] |
KLA | 1.01 |
1.07 |
1.03 |
1.03 |
1.04 |
1.00 |
1.06 |
| ATP | .99 |
1.09 |
1.08 |
1.05 |
1.02 |
1.06 |
1.07 |
| KLA/ATP | 1.00 |
.90 |
1.06 |
1.14 |
1.10 |
1.08 |
1.09 |
|
| Ptprc | 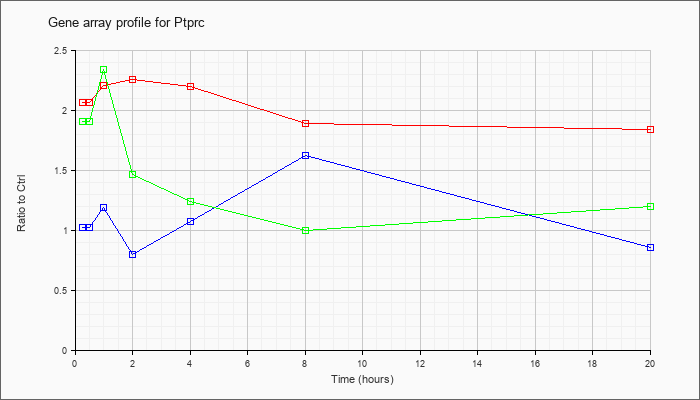 |
AK088215 |
2 days neonate thymus thymic cells cDNA, RIKEN full-length enriched library, clone:E430007G18 product:protein tyrosine phosphatase, receptor type, C, full insert sequence. [AK088215] |
KLA | 2.15 |
1.93 |
2.11 |
1.83 |
2.08 |
1.72 |
1.51 |
| ATP | .95 |
.84 |
.65 |
.50 |
1.79 |
.82 |
.77 |
| KLA/ATP | 2.01 |
1.68 |
1.50 |
.80 |
1.16 |
.73 |
1.14 |
|
| Ptprc |  |
NM_011210 |
protein tyrosine phosphatase, receptor type, C (Ptprc), transcript variant 2, mRNA [NM_011210] |
KLA | 2.06 |
2.00 |
2.21 |
2.29 |
2.20 |
1.90 |
1.86 |
| ATP | 1.02 |
.99 |
1.23 |
.82 |
1.01 |
1.68 |
.86 |
| KLA/ATP | 1.89 |
2.00 |
2.40 |
1.52 |
1.24 |
1.01 |
1.20 |
|
| Rac1 | 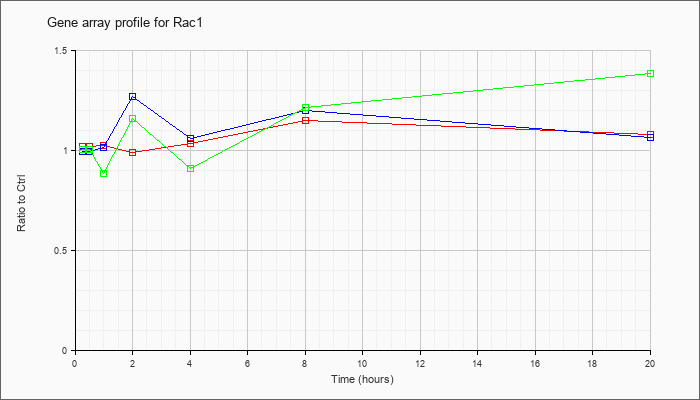 |
NM_009007 |
RAS-related C3 botulinum substrate 1 (Rac1), mRNA [NM_009007] |
KLA | 1.02 |
1.01 |
1.02 |
.99 |
1.03 |
1.15 |
1.08 |
| ATP | .99 |
1.15 |
1.01 |
1.27 |
1.06 |
1.20 |
1.06 |
| KLA/ATP | 1.00 |
1.04 |
.88 |
1.16 |
.91 |
1.21 |
1.38 |
|
| Rac2 | 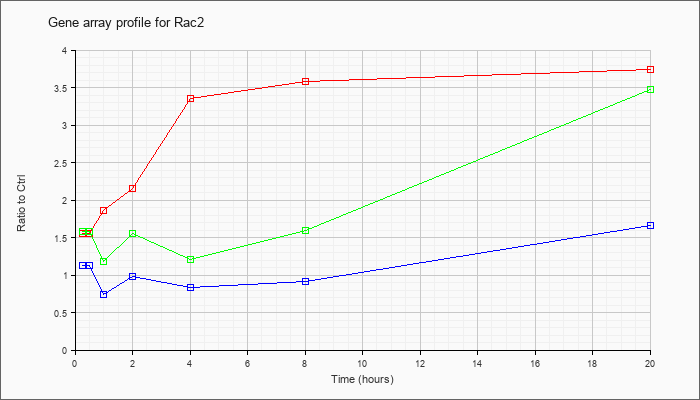 |
NM_009008 |
RAS-related C3 botulinum substrate 2 (Rac2), mRNA [NM_009008] |
KLA | 1.55 |
1.68 |
1.86 |
2.15 |
3.36 |
3.59 |
3.74 |
| ATP | 1.13 |
1.16 |
.74 |
.99 |
.84 |
.91 |
1.66 |
| KLA/ATP | 1.59 |
1.71 |
1.19 |
1.56 |
1.21 |
1.60 |
3.48 |
|
| Raf1 | 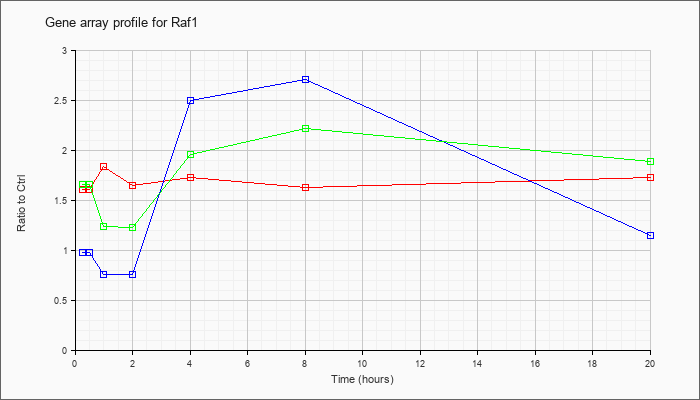 |
AK036317 |
16 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:9630056E02 product:v-raf-1 leukemia viral oncogene 1, full insert sequence. [AK036317] |
KLA | 1.47 |
1.34 |
1.55 |
1.29 |
1.59 |
1.32 |
1.46 |
| ATP | 1.05 |
.98 |
.54 |
.63 |
3.32 |
2.07 |
1.01 |
| KLA/ATP | 1.57 |
1.33 |
.79 |
.96 |
2.26 |
1.38 |
1.35 |
|
| Raf1 |  |
NM_029780 |
v-raf-leukemia viral oncogene 1 (Raf1), mRNA [NM_029780] |
KLA | 1.65 |
1.64 |
1.93 |
1.77 |
1.77 |
1.72 |
1.81 |
| ATP | .95 |
.96 |
.83 |
.80 |
2.22 |
2.92 |
1.19 |
| KLA/ATP | 1.68 |
1.64 |
1.39 |
1.32 |
1.85 |
2.50 |
2.06 |
|
| Rps6kb1 | 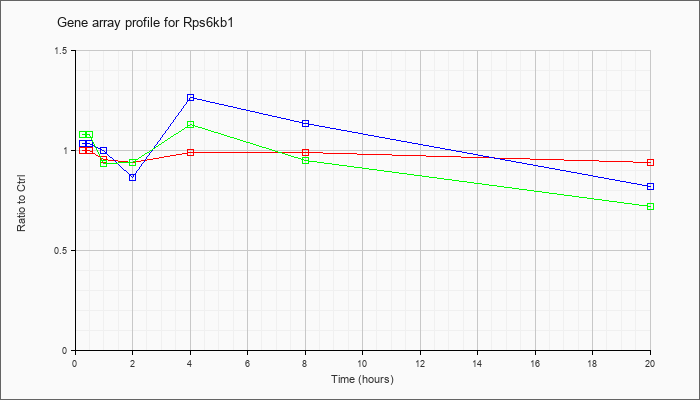 |
NM_001114334 |
ribosomal protein S6 kinase, polypeptide 1 (Rps6kb1), transcript variant 1, mRNA [NM_001114334] |
KLA | .99 |
1.00 |
.90 |
.89 |
.99 |
.99 |
.92 |
| ATP | 1.01 |
.96 |
1.00 |
.79 |
1.36 |
1.19 |
.72 |
| KLA/ATP | 1.08 |
.99 |
.93 |
.92 |
1.18 |
.90 |
.56 |
|
| Rps6kb1 |  |
NM_028259 |
ribosomal protein S6 kinase, polypeptide 1 (Rps6kb1), transcript variant 2, mRNA [NM_028259] |
KLA | 1.01 |
.93 |
1.07 |
1.04 |
.99 |
.99 |
.98 |
| ATP | 1.09 |
1.05 |
1.00 |
1.01 |
1.08 |
1.03 |
1.03 |
| KLA/ATP | 1.07 |
1.04 |
.94 |
.98 |
1.03 |
1.05 |
1.05 |
|
| Rps6kb2 | 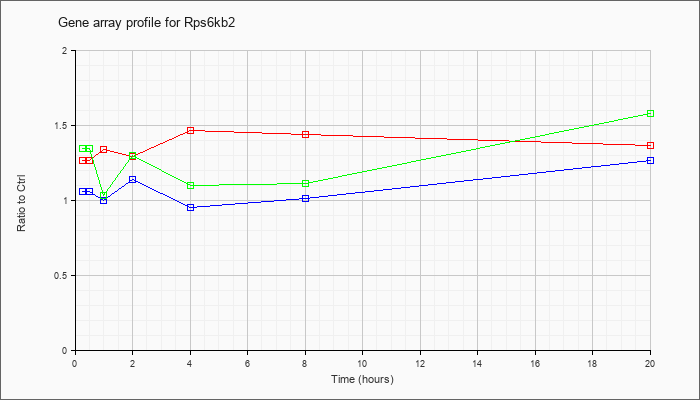 |
AK090093 |
kidney CCL-142 RAG cDNA, RIKEN full-length enriched library, clone:G430107K15 product:ribosomal protein S6 kinase, 70kD, polypeptide 2, full insert sequence [AK090093] |
KLA | .97 |
1.01 |
.98 |
.97 |
.98 |
1.04 |
1.07 |
| ATP | 1.00 |
1.08 |
1.06 |
1.09 |
1.11 |
1.06 |
1.03 |
| KLA/ATP | 1.05 |
.96 |
.86 |
1.03 |
1.40 |
1.07 |
.99 |
|
| Rps6kb2 |  |
NM_021485 |
ribosomal protein S6 kinase, polypeptide 2 (Rps6kb2), mRNA [NM_021485] |
KLA | 1.36 |
1.40 |
1.45 |
1.40 |
1.63 |
1.57 |
1.46 |
| ATP | 1.08 |
1.19 |
.97 |
1.15 |
.90 |
.99 |
1.34 |
| KLA/ATP | 1.44 |
1.55 |
1.09 |
1.39 |
1.00 |
1.12 |
1.77 |
|
| Scin | 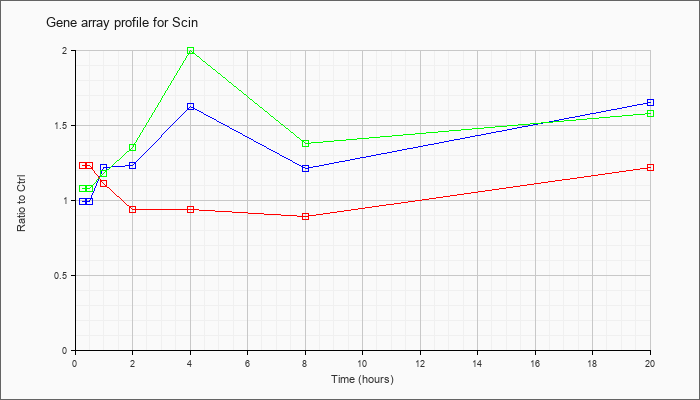 |
NM_009132 |
scinderin (Scin), mRNA [NM_009132] |
KLA | 1.23 |
1.20 |
1.11 |
.94 |
.94 |
.89 |
1.22 |
| ATP | .99 |
.96 |
1.22 |
1.23 |
1.62 |
1.21 |
1.65 |
| KLA/ATP | 1.08 |
1.26 |
1.18 |
1.35 |
2.00 |
1.38 |
1.58 |
|
| Sphk1 | 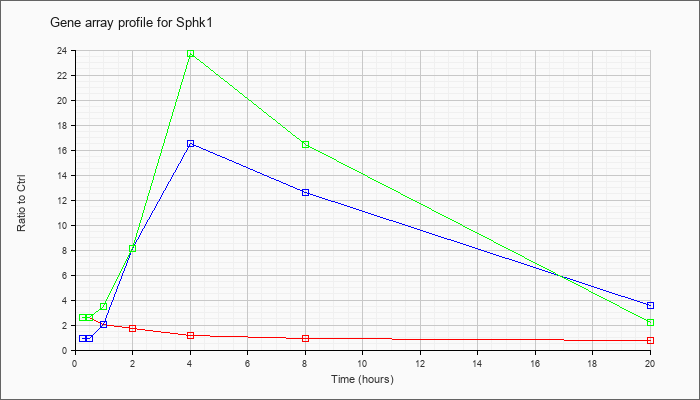 |
NM_025367 |
sphingosine kinase 1 (Sphk1), transcript variant 2, mRNA [NM_025367] |
KLA | 2.61 |
2.50 |
2.04 |
1.71 |
1.19 |
.91 |
.74 |
| ATP | .94 |
1.00 |
2.06 |
8.09 |
16.52 |
12.59 |
3.53 |
| KLA/ATP | 2.64 |
2.81 |
3.50 |
8.13 |
23.71 |
16.46 |
2.22 |
|
| Sphk2 | 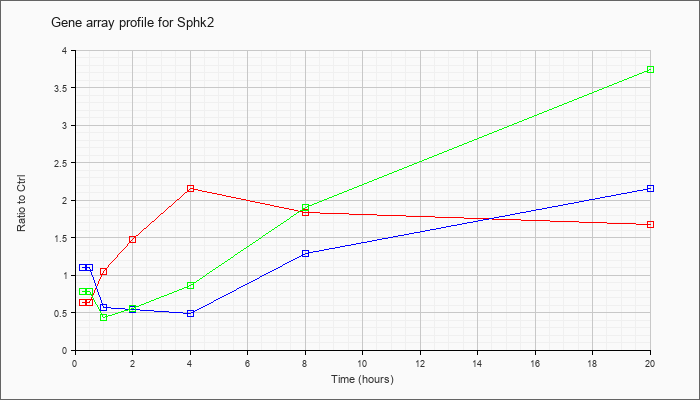 |
NM_020011 |
sphingosine kinase 2 (Sphk2), transcript variant 2, mRNA [NM_020011] |
KLA | .64 |
.74 |
1.05 |
1.47 |
2.15 |
1.84 |
1.67 |
| ATP | 1.10 |
1.02 |
.57 |
.54 |
.49 |
1.29 |
2.15 |
| KLA/ATP | .78 |
.64 |
.43 |
.56 |
.86 |
1.90 |
3.74 |
|
| Syk | 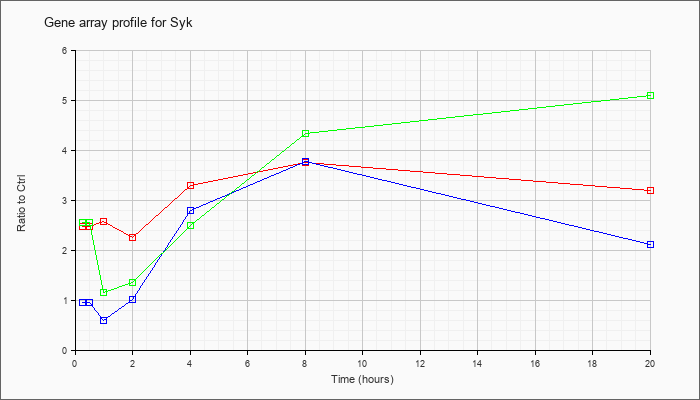 |
NM_011518 |
spleen tyrosine kinase (Syk), mRNA [NM_011518] |
KLA | 2.48 |
2.59 |
2.57 |
2.26 |
3.29 |
3.75 |
3.19 |
| ATP | .96 |
.85 |
.59 |
1.01 |
2.80 |
3.77 |
2.11 |
| KLA/ATP | 2.56 |
2.09 |
1.16 |
1.35 |
2.50 |
4.33 |
5.10 |
|
| Vasp | 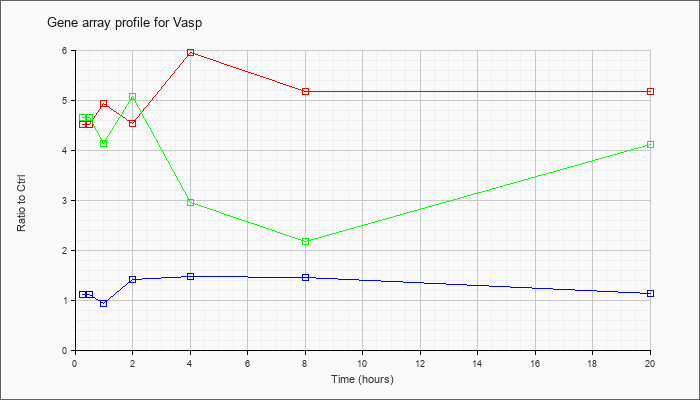 |
NM_009499 |
vasodilator-stimulated phosphoprotein (Vasp), mRNA [NM_009499] |
KLA | 4.52 |
4.78 |
4.94 |
4.54 |
5.95 |
5.17 |
5.17 |
| ATP | 1.11 |
1.31 |
.93 |
1.41 |
1.48 |
1.45 |
1.14 |
| KLA/ATP | 4.65 |
5.51 |
4.13 |
5.08 |
2.96 |
2.18 |
4.11 |
|
| Vav1 | 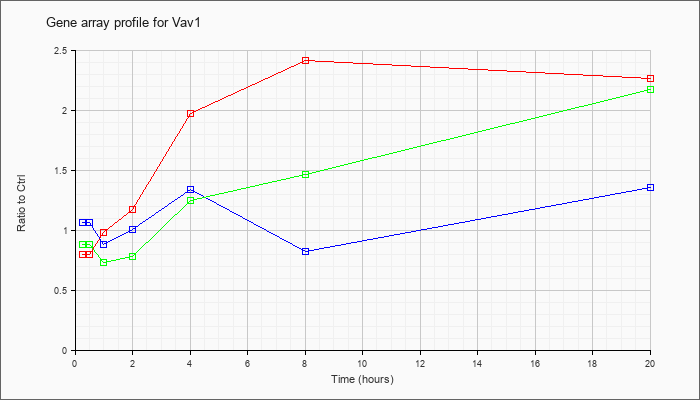 |
NM_011691 |
vav 1 oncogene (Vav1), mRNA [NM_011691] |
KLA | .79 |
.83 |
.98 |
1.17 |
1.97 |
2.42 |
2.26 |
| ATP | 1.06 |
1.02 |
.88 |
1.01 |
1.34 |
.82 |
1.36 |
| KLA/ATP | .88 |
.85 |
.73 |
.78 |
1.25 |
1.47 |
2.17 |
|
| Vav2 | 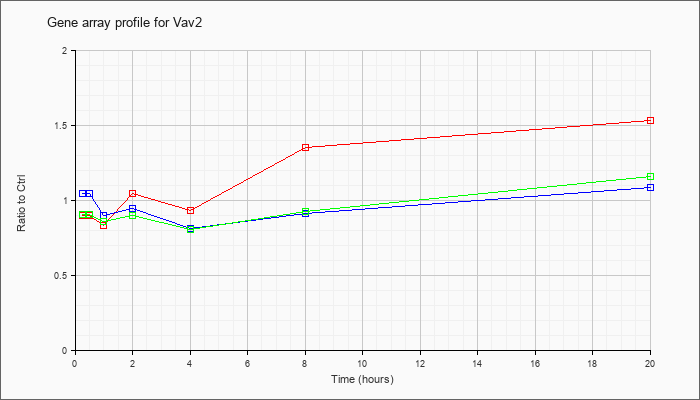 |
BC053060 |
vav 2 oncogene, mRNA (cDNA clone MGC:62363 IMAGE:5704852), complete cds [BC053060] |
KLA | .73 |
.58 |
.55 |
1.16 |
.76 |
1.35 |
1.75 |
| ATP | .99 |
.89 |
.79 |
.82 |
.67 |
.85 |
1.09 |
| KLA/ATP | .64 |
.63 |
.61 |
.65 |
.66 |
.80 |
1.23 |
|
| Vav2 |  |
NM_009500 |
vav 2 oncogene (Vav2), mRNA [NM_009500] |
KLA | .98 |
1.05 |
.98 |
.99 |
1.02 |
1.36 |
1.43 |
| ATP | 1.07 |
1.06 |
.96 |
1.01 |
.89 |
.95 |
1.08 |
| KLA/ATP | 1.04 |
1.05 |
.98 |
1.03 |
.88 |
.99 |
1.13 |
|
| Vav3 | 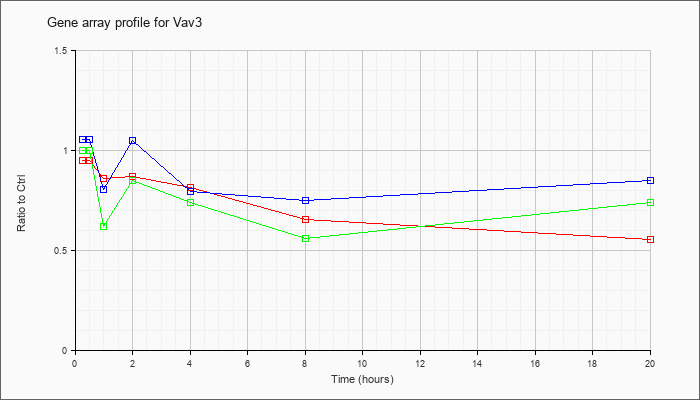 |
NM_020505 |
vav 3 oncogene (Vav3), transcript variant 1, mRNA [NM_020505] |
KLA | .95 |
.94 |
.86 |
.87 |
.82 |
.66 |
.56 |
| ATP | 1.06 |
1.09 |
.81 |
1.05 |
.80 |
.75 |
.85 |
| KLA/ATP | 1.00 |
.84 |
.62 |
.85 |
.74 |
.56 |
.74 |
|
| Was | 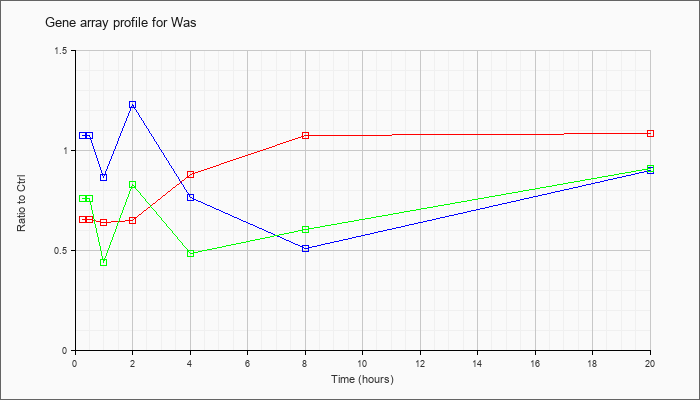 |
NM_009515 |
Wiskott-Aldrich syndrome homolog (human) (Was), mRNA [NM_009515] |
KLA | .66 |
.67 |
.64 |
.65 |
.88 |
1.08 |
1.09 |
| ATP | 1.08 |
1.11 |
.87 |
1.23 |
.77 |
.51 |
.90 |
| KLA/ATP | .76 |
.70 |
.44 |
.83 |
.49 |
.61 |
.91 |
|
| Wasf1 |  |
NM_031877 |
WASP family 1 (Wasf1), mRNA [NM_031877] |
KLA | .75 |
.69 |
.77 |
.81 |
.69 |
.66 |
.54 |
| ATP | 1.01 |
.97 |
.93 |
.98 |
.70 |
.66 |
.97 |
| KLA/ATP | .75 |
.73 |
.80 |
.79 |
.73 |
.67 |
.56 |
|
| Wasf2 | 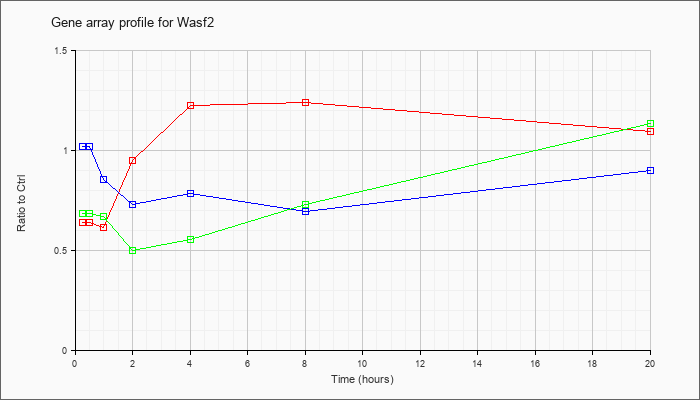 |
AK150140 |
bone marrow macrophage cDNA, RIKEN full-length enriched library, clone:G530136J12 product:WAS protein family, member 2, full insert sequence [AK150140] |
KLA | .58 |
.52 |
.52 |
.83 |
1.06 |
1.17 |
.91 |
| ATP | .97 |
.89 |
.93 |
.57 |
.72 |
.72 |
.86 |
| KLA/ATP | .55 |
.45 |
.56 |
.36 |
.45 |
.65 |
1.08 |
|
| Wasf2 |  |
NM_153423 |
WAS protein family, member 2 (Wasf2), mRNA [NM_153423] |
KLA | .67 |
.75 |
.66 |
1.01 |
1.31 |
1.28 |
1.19 |
| ATP | 1.05 |
1.10 |
.82 |
.81 |
.82 |
.68 |
.92 |
| KLA/ATP | .75 |
.76 |
.73 |
.57 |
.61 |
.77 |
1.16 |
|
| Wasf3 | 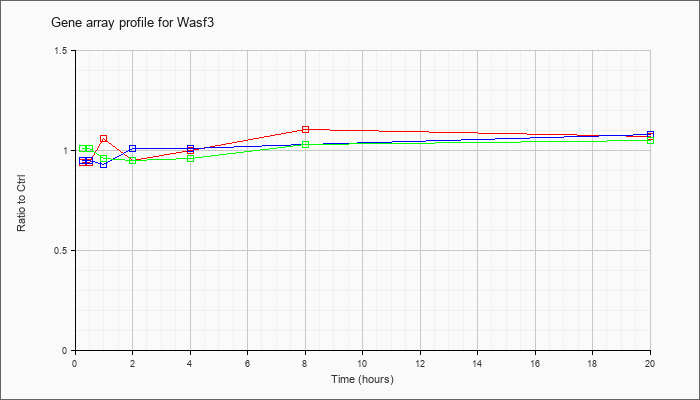 |
NM_145155 |
WAS protein family, member 3 (Wasf3), mRNA [NM_145155] |
KLA | .94 |
.95 |
1.06 |
.95 |
1.00 |
1.10 |
1.07 |
| ATP | .95 |
.96 |
.93 |
1.01 |
1.01 |
1.03 |
1.08 |
| KLA/ATP | 1.01 |
.91 |
.96 |
.95 |
.96 |
1.03 |
1.05 |
|
| Wasl | 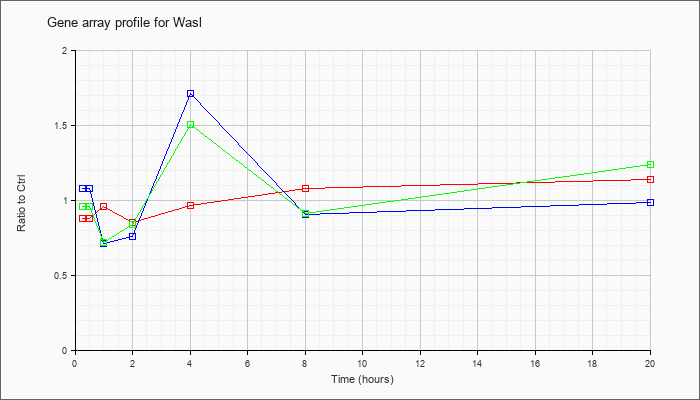 |
NM_028459 |
Wiskott-Aldrich syndrome-like (human) (Wasl), mRNA [NM_028459] |
KLA | .88 |
.89 |
.96 |
.85 |
.97 |
1.08 |
1.14 |
| ATP | 1.08 |
.99 |
.71 |
.76 |
1.71 |
.91 |
.99 |
| KLA/ATP | .96 |
.84 |
.72 |
.84 |
1.51 |
.91 |
1.24 |
|